DIFFUSE LARGE B-CELL LYMPHOMA (DLBCL) STUDY REVEALS BIOLOGICAL HETEROGENEITY BETWEEN ETHNICALLY DIVERSE COUNTRIES; VALIDATION OF 6-GENE PROGNOSTIC SCORE IN AN INTERNATIONAL COHORT
(Abstract release date: 05/19/16)
EHA Library. Tekin-Bubenheim N. 06/09/16; 132950; E1401
Disclosure(s): R.A. Padua receives research support from Abbvie and Genetech.

Dr. Nilgun Tekin-Bubenheim
Contributions
Contributions
Abstract
Abstract: E1401
Type: Eposter Presentation
Background
Biological variation may arise from ethnic diversity or environment which may influence disease or host response. The International Atomic Energy Agency sponsored a prospective cohort study of diffuse large B-cell lymphoma (DLBCL) in countries from 5 United Nations-defined geographical regions to test this hypothesis.
Aims
The molecular analysis of DLBCL was conducted aiming to use the previously reported 6-gene score (LMO2, BCL6, FN1, CCND2, SCYA3 and BCL2) (Lossos N Engl J Med 2004; 350: 1828–37) as a predictor of prognosis in DLBCL and validation of the utility of this scoring system in an international cohort.
Methods
Consented patients with DLBCL (n=162) in Hungary (n=28), Chile (n=27), India (n=27), Thailand (n=27), Brazil (n=18), Turkey (n=15), Philippines (n=13) and S Korea (n=7) were treated with R-CHOP between 2008 and 2013. RNA from formalin fixed paraffin embedded (FFPE) diagnostic tissue was shipped to a central laboratory. Expression of the 6 genes were assayed, using either high volume or low volume (Brazil) cDNA, by Taqman QPCR and relative quantification assigned based on expression ratio to normalised copy number. In a multivariate analysis, as described in the following equation: mortality-predictor score = (-0.0273xLMO2) + (-0.2103xBCL6) + (-0.1878xFN1) + (0.0346xCCND2) + (0.1888xSCYA3) + (0.5527xBCL2), variation in gene expression by country was investigated using analysis of variance, while variation in event-free (EFS) and overall survival (OS) for the whole cohort was investigated using Cox proportional-hazards regression models.
Results
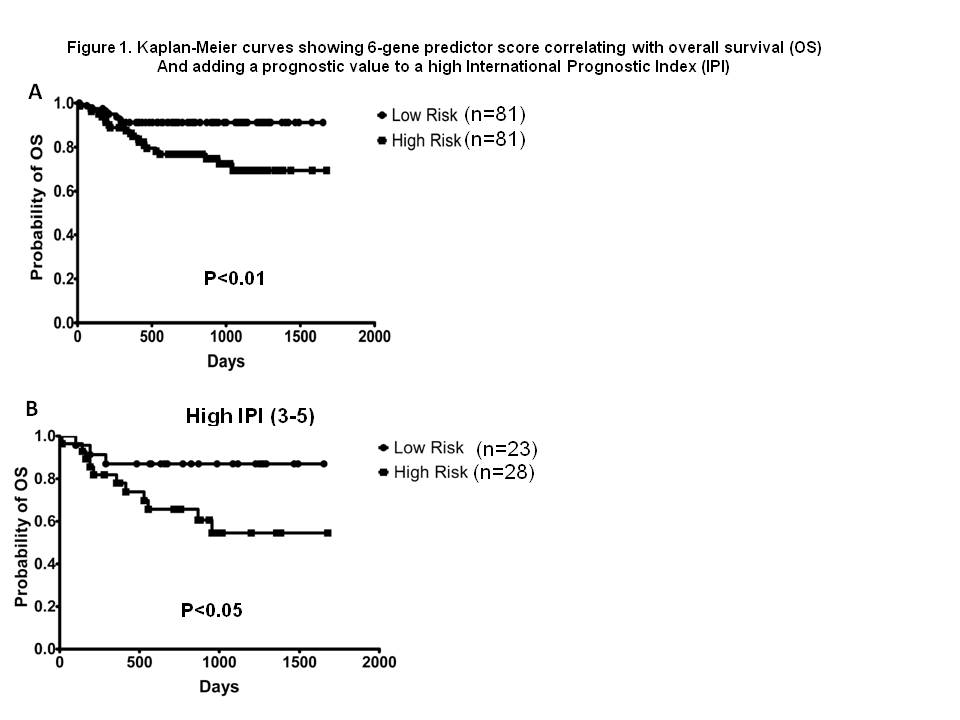
There was significant inter-country variation for all 6 genes individually (p<0.0001), and when combined in the 6-gene model (p<0.0001). The variation in 2y EFS between countries ranged from 56% (Turkey) to 85% (Chile) With the analysis of the cohort as a whole the 6-gene score returned a hazard ratio of 0.35 (95% CI 0.17–0.74) for OS, demonstrating the score can stratify relative survival risk. To further develop the prognostic value, patient mortality-predictor scores were ranked into 2 groups with lower or higher risk of death. In a univariate Kaplan–Meier model of these 2 groups [low (n=81), and high (n=81)], defined by the prediction model based on the weighted expression of the 6 genes (Malumbres Blood 2008; 111:5509-5514), there was no significant difference in EFS between low and high risk groups (p=0.18); however, a significant difference in OS between low and high risk (p<0.01) was observed (Figure 1A). When patients with a high international prognostic indicator (IPI) (3-5), n=51, and low IPI (0-2), n=111 were analysed separately, the 6-gene score did not add prognostic value to the low IPI cases (p=0.08), but did add additional predictive value for OS, though not EFS, in high IPI cases (Figure 1B, p<0.05). There was no significant difference between patients treated with CHOP (n=28) or R-CHOP (n=132) for EFS or OS. Conclusion: The analysis of this cohort showed that the 6-gene model added prognostic information for overall survival in high IPI cases, thereby validating its utility for predicting outcome in an international setting.
Conclusion
The analysis of this cohort showed that the 6-gene model added prognostic information for overall survival in high IPI cases, thereby validating its utility for predicting outcome in an international setting.
Session topic: E-poster
Keyword(s): DLBCL, Gene expression, Prognosis
Type: Eposter Presentation
Background
Biological variation may arise from ethnic diversity or environment which may influence disease or host response. The International Atomic Energy Agency sponsored a prospective cohort study of diffuse large B-cell lymphoma (DLBCL) in countries from 5 United Nations-defined geographical regions to test this hypothesis.
Aims
The molecular analysis of DLBCL was conducted aiming to use the previously reported 6-gene score (LMO2, BCL6, FN1, CCND2, SCYA3 and BCL2) (Lossos N Engl J Med 2004; 350: 1828–37) as a predictor of prognosis in DLBCL and validation of the utility of this scoring system in an international cohort.
Methods
Consented patients with DLBCL (n=162) in Hungary (n=28), Chile (n=27), India (n=27), Thailand (n=27), Brazil (n=18), Turkey (n=15), Philippines (n=13) and S Korea (n=7) were treated with R-CHOP between 2008 and 2013. RNA from formalin fixed paraffin embedded (FFPE) diagnostic tissue was shipped to a central laboratory. Expression of the 6 genes were assayed, using either high volume or low volume (Brazil) cDNA, by Taqman QPCR and relative quantification assigned based on expression ratio to normalised copy number. In a multivariate analysis, as described in the following equation: mortality-predictor score = (-0.0273xLMO2) + (-0.2103xBCL6) + (-0.1878xFN1) + (0.0346xCCND2) + (0.1888xSCYA3) + (0.5527xBCL2), variation in gene expression by country was investigated using analysis of variance, while variation in event-free (EFS) and overall survival (OS) for the whole cohort was investigated using Cox proportional-hazards regression models.
Results
There was significant inter-country variation for all 6 genes individually (p<0.0001), and when combined in the 6-gene model (p<0.0001). The variation in 2y EFS between countries ranged from 56% (Turkey) to 85% (Chile) With the analysis of the cohort as a whole the 6-gene score returned a hazard ratio of 0.35 (95% CI 0.17–0.74) for OS, demonstrating the score can stratify relative survival risk. To further develop the prognostic value, patient mortality-predictor scores were ranked into 2 groups with lower or higher risk of death. In a univariate Kaplan–Meier model of these 2 groups [low (n=81), and high (n=81)], defined by the prediction model based on the weighted expression of the 6 genes (Malumbres Blood 2008; 111:5509-5514), there was no significant difference in EFS between low and high risk groups (p=0.18); however, a significant difference in OS between low and high risk (p<0.01) was observed (Figure 1A). When patients with a high international prognostic indicator (IPI) (3-5), n=51, and low IPI (0-2), n=111 were analysed separately, the 6-gene score did not add prognostic value to the low IPI cases (p=0.08), but did add additional predictive value for OS, though not EFS, in high IPI cases (Figure 1B, p<0.05). There was no significant difference between patients treated with CHOP (n=28) or R-CHOP (n=132) for EFS or OS. Conclusion: The analysis of this cohort showed that the 6-gene model added prognostic information for overall survival in high IPI cases, thereby validating its utility for predicting outcome in an international setting.
Conclusion
The analysis of this cohort showed that the 6-gene model added prognostic information for overall survival in high IPI cases, thereby validating its utility for predicting outcome in an international setting.

Session topic: E-poster
Keyword(s): DLBCL, Gene expression, Prognosis
Abstract: E1401
Type: Eposter Presentation
Background
Biological variation may arise from ethnic diversity or environment which may influence disease or host response. The International Atomic Energy Agency sponsored a prospective cohort study of diffuse large B-cell lymphoma (DLBCL) in countries from 5 United Nations-defined geographical regions to test this hypothesis.
Aims
The molecular analysis of DLBCL was conducted aiming to use the previously reported 6-gene score (LMO2, BCL6, FN1, CCND2, SCYA3 and BCL2) (Lossos N Engl J Med 2004; 350: 1828–37) as a predictor of prognosis in DLBCL and validation of the utility of this scoring system in an international cohort.
Methods
Consented patients with DLBCL (n=162) in Hungary (n=28), Chile (n=27), India (n=27), Thailand (n=27), Brazil (n=18), Turkey (n=15), Philippines (n=13) and S Korea (n=7) were treated with R-CHOP between 2008 and 2013. RNA from formalin fixed paraffin embedded (FFPE) diagnostic tissue was shipped to a central laboratory. Expression of the 6 genes were assayed, using either high volume or low volume (Brazil) cDNA, by Taqman QPCR and relative quantification assigned based on expression ratio to normalised copy number. In a multivariate analysis, as described in the following equation: mortality-predictor score = (-0.0273xLMO2) + (-0.2103xBCL6) + (-0.1878xFN1) + (0.0346xCCND2) + (0.1888xSCYA3) + (0.5527xBCL2), variation in gene expression by country was investigated using analysis of variance, while variation in event-free (EFS) and overall survival (OS) for the whole cohort was investigated using Cox proportional-hazards regression models.
Results
There was significant inter-country variation for all 6 genes individually (p<0.0001), and when combined in the 6-gene model (p<0.0001). The variation in 2y EFS between countries ranged from 56% (Turkey) to 85% (Chile) With the analysis of the cohort as a whole the 6-gene score returned a hazard ratio of 0.35 (95% CI 0.17–0.74) for OS, demonstrating the score can stratify relative survival risk. To further develop the prognostic value, patient mortality-predictor scores were ranked into 2 groups with lower or higher risk of death. In a univariate Kaplan–Meier model of these 2 groups [low (n=81), and high (n=81)], defined by the prediction model based on the weighted expression of the 6 genes (Malumbres Blood 2008; 111:5509-5514), there was no significant difference in EFS between low and high risk groups (p=0.18); however, a significant difference in OS between low and high risk (p<0.01) was observed (Figure 1A). When patients with a high international prognostic indicator (IPI) (3-5), n=51, and low IPI (0-2), n=111 were analysed separately, the 6-gene score did not add prognostic value to the low IPI cases (p=0.08), but did add additional predictive value for OS, though not EFS, in high IPI cases (Figure 1B, p<0.05). There was no significant difference between patients treated with CHOP (n=28) or R-CHOP (n=132) for EFS or OS. Conclusion: The analysis of this cohort showed that the 6-gene model added prognostic information for overall survival in high IPI cases, thereby validating its utility for predicting outcome in an international setting.
Conclusion
The analysis of this cohort showed that the 6-gene model added prognostic information for overall survival in high IPI cases, thereby validating its utility for predicting outcome in an international setting.

Session topic: E-poster
Keyword(s): DLBCL, Gene expression, Prognosis
Type: Eposter Presentation
Background
Biological variation may arise from ethnic diversity or environment which may influence disease or host response. The International Atomic Energy Agency sponsored a prospective cohort study of diffuse large B-cell lymphoma (DLBCL) in countries from 5 United Nations-defined geographical regions to test this hypothesis.
Aims
The molecular analysis of DLBCL was conducted aiming to use the previously reported 6-gene score (LMO2, BCL6, FN1, CCND2, SCYA3 and BCL2) (Lossos N Engl J Med 2004; 350: 1828–37) as a predictor of prognosis in DLBCL and validation of the utility of this scoring system in an international cohort.
Methods
Consented patients with DLBCL (n=162) in Hungary (n=28), Chile (n=27), India (n=27), Thailand (n=27), Brazil (n=18), Turkey (n=15), Philippines (n=13) and S Korea (n=7) were treated with R-CHOP between 2008 and 2013. RNA from formalin fixed paraffin embedded (FFPE) diagnostic tissue was shipped to a central laboratory. Expression of the 6 genes were assayed, using either high volume or low volume (Brazil) cDNA, by Taqman QPCR and relative quantification assigned based on expression ratio to normalised copy number. In a multivariate analysis, as described in the following equation: mortality-predictor score = (-0.0273xLMO2) + (-0.2103xBCL6) + (-0.1878xFN1) + (0.0346xCCND2) + (0.1888xSCYA3) + (0.5527xBCL2), variation in gene expression by country was investigated using analysis of variance, while variation in event-free (EFS) and overall survival (OS) for the whole cohort was investigated using Cox proportional-hazards regression models.
Results
There was significant inter-country variation for all 6 genes individually (p<0.0001), and when combined in the 6-gene model (p<0.0001). The variation in 2y EFS between countries ranged from 56% (Turkey) to 85% (Chile) With the analysis of the cohort as a whole the 6-gene score returned a hazard ratio of 0.35 (95% CI 0.17–0.74) for OS, demonstrating the score can stratify relative survival risk. To further develop the prognostic value, patient mortality-predictor scores were ranked into 2 groups with lower or higher risk of death. In a univariate Kaplan–Meier model of these 2 groups [low (n=81), and high (n=81)], defined by the prediction model based on the weighted expression of the 6 genes (Malumbres Blood 2008; 111:5509-5514), there was no significant difference in EFS between low and high risk groups (p=0.18); however, a significant difference in OS between low and high risk (p<0.01) was observed (Figure 1A). When patients with a high international prognostic indicator (IPI) (3-5), n=51, and low IPI (0-2), n=111 were analysed separately, the 6-gene score did not add prognostic value to the low IPI cases (p=0.08), but did add additional predictive value for OS, though not EFS, in high IPI cases (Figure 1B, p<0.05). There was no significant difference between patients treated with CHOP (n=28) or R-CHOP (n=132) for EFS or OS. Conclusion: The analysis of this cohort showed that the 6-gene model added prognostic information for overall survival in high IPI cases, thereby validating its utility for predicting outcome in an international setting.
Conclusion
The analysis of this cohort showed that the 6-gene model added prognostic information for overall survival in high IPI cases, thereby validating its utility for predicting outcome in an international setting.

Session topic: E-poster
Keyword(s): DLBCL, Gene expression, Prognosis
{{ help_message }}
{{filter}}


