SEARCHING FOR NOVEL PROGNOSTIC BIOMARKERS IN DIFFUSE LARGE B CELL LYMPHOMA (DLBCL) BY CIBERSORT-BASED ANALYSIS OF TUMOR MICROENVIRONMENT.
(Abstract release date: 05/19/16)
EHA Library. Ciavarella S. 06/09/16; 132924; E1375

Dr. Sabino Ciavarella
Contributions
Contributions
Abstract
Abstract: E1375
Type: Eposter Presentation
Background
Diffuse large B cell lymphoma (DLBCL) comprises a large group of disease entities with high molecular heterogeneity and variable treatment responsiveness. Results from gene expression profiling (GEP) studies highlighted the role of cell of origin, namely activated B cell-like cells (ABC) and germinal center B cells (GCB), and stromal gene signatures for predicting clinical outcome and stratifying patient risk. However, translation of GEP information in easily applicable prognostic biomarkers remains a challenge.
Aims
We applied an innovative GEP-based computational method, namely CIBERSORT, to determine the cellular composition of DLBCL microenvironment and investigate associations between abundance of certain tumor-infiltrating non-malignant cell types and molecular/clinical traits of the disease.
Methods
We generated a gene signature matrix including a total number of 1028 genes that distinguish 17 different immune and non-immune cell types, the latter including adipocytes, endothelial cells (EC), pericytes and myofibroblasts (MF). This matrix was used to perform CIBERSORT analysis (on-line access to the webserver http://cibersort.stanford.edu/) of GEP data from 3 publicly datasets (GSE10846, GSE19246 and GSE34171) of overall 604 DLBCL cases. The relative percentage of each tumor-infiltrating cell fraction was estimated and a global heatmap of tumor microenvironment composition was generated. Results were also stratified according to either cell of origin or clinical outcome data and significant differences in each subgroup calculated by a two-sided Mann-Whitney test.
Results
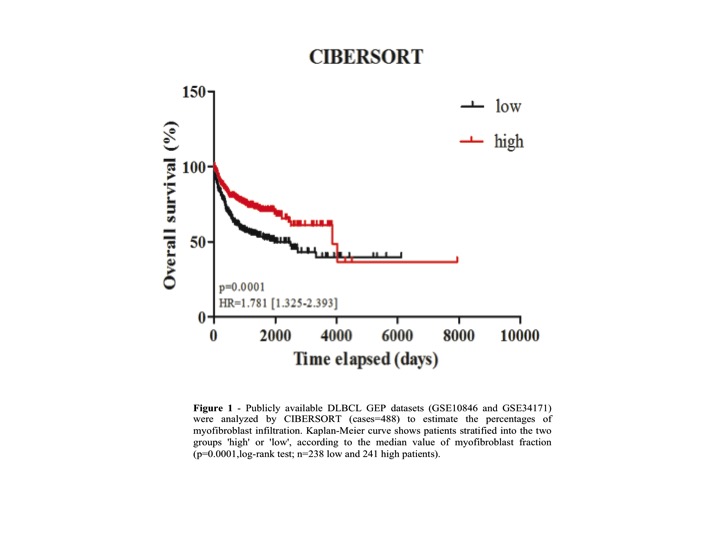
By applying CIBERSORT, we found that, among immune cells, memory B cells, plasma cells, NK cells, dendritic cells, CD4 T cells and macrophages mainly infiltrate DLBCL tissues. In particular, plasma cells, NK and dendritic cells showed significant higher proportion in ABC than GCB tumors, whereas CD4 T-cells and dendritic cells were prevalent in patients with better outcome. Notably, among stromal cells, MF was the most represented population and exhibited a significant predominance in patients with more favorable outcome and a strong positive correlation with overall survival (Figure 1). Consistently, the fraction of MF was significantly larger in GCB tumors compared with ABC subtypes. These latter resulted more enriched in EC.
Conclusion
By estimating relative fractions of DLBCL-infiltrating cells using CIBERSORT, we studied unknown aspects of tumor microenvironment with potential prognostic implications. Our data suggest that the composition of DLBCL with diverse molecular and clinical features also differ in term of immune and, particularly, stromal cell infiltration. These findings are in line with previous reports highlighting the prognostic value of specific stromal gene signatures and support further studies aimed at uncover novel prognostic/predictive biomarkers correlated with non-malignant tumor-associated cell subsets.
Session topic: E-poster
Keyword(s): Diffuse large B cell lymphoma, Microenvironment, Prognostic factor, Stromal cell
Type: Eposter Presentation
Background
Diffuse large B cell lymphoma (DLBCL) comprises a large group of disease entities with high molecular heterogeneity and variable treatment responsiveness. Results from gene expression profiling (GEP) studies highlighted the role of cell of origin, namely activated B cell-like cells (ABC) and germinal center B cells (GCB), and stromal gene signatures for predicting clinical outcome and stratifying patient risk. However, translation of GEP information in easily applicable prognostic biomarkers remains a challenge.
Aims
We applied an innovative GEP-based computational method, namely CIBERSORT, to determine the cellular composition of DLBCL microenvironment and investigate associations between abundance of certain tumor-infiltrating non-malignant cell types and molecular/clinical traits of the disease.
Methods
We generated a gene signature matrix including a total number of 1028 genes that distinguish 17 different immune and non-immune cell types, the latter including adipocytes, endothelial cells (EC), pericytes and myofibroblasts (MF). This matrix was used to perform CIBERSORT analysis (on-line access to the webserver http://cibersort.stanford.edu/) of GEP data from 3 publicly datasets (GSE10846, GSE19246 and GSE34171) of overall 604 DLBCL cases. The relative percentage of each tumor-infiltrating cell fraction was estimated and a global heatmap of tumor microenvironment composition was generated. Results were also stratified according to either cell of origin or clinical outcome data and significant differences in each subgroup calculated by a two-sided Mann-Whitney test.
Results
By applying CIBERSORT, we found that, among immune cells, memory B cells, plasma cells, NK cells, dendritic cells, CD4 T cells and macrophages mainly infiltrate DLBCL tissues. In particular, plasma cells, NK and dendritic cells showed significant higher proportion in ABC than GCB tumors, whereas CD4 T-cells and dendritic cells were prevalent in patients with better outcome. Notably, among stromal cells, MF was the most represented population and exhibited a significant predominance in patients with more favorable outcome and a strong positive correlation with overall survival (Figure 1). Consistently, the fraction of MF was significantly larger in GCB tumors compared with ABC subtypes. These latter resulted more enriched in EC.
Conclusion
By estimating relative fractions of DLBCL-infiltrating cells using CIBERSORT, we studied unknown aspects of tumor microenvironment with potential prognostic implications. Our data suggest that the composition of DLBCL with diverse molecular and clinical features also differ in term of immune and, particularly, stromal cell infiltration. These findings are in line with previous reports highlighting the prognostic value of specific stromal gene signatures and support further studies aimed at uncover novel prognostic/predictive biomarkers correlated with non-malignant tumor-associated cell subsets.

Session topic: E-poster
Keyword(s): Diffuse large B cell lymphoma, Microenvironment, Prognostic factor, Stromal cell
Abstract: E1375
Type: Eposter Presentation
Background
Diffuse large B cell lymphoma (DLBCL) comprises a large group of disease entities with high molecular heterogeneity and variable treatment responsiveness. Results from gene expression profiling (GEP) studies highlighted the role of cell of origin, namely activated B cell-like cells (ABC) and germinal center B cells (GCB), and stromal gene signatures for predicting clinical outcome and stratifying patient risk. However, translation of GEP information in easily applicable prognostic biomarkers remains a challenge.
Aims
We applied an innovative GEP-based computational method, namely CIBERSORT, to determine the cellular composition of DLBCL microenvironment and investigate associations between abundance of certain tumor-infiltrating non-malignant cell types and molecular/clinical traits of the disease.
Methods
We generated a gene signature matrix including a total number of 1028 genes that distinguish 17 different immune and non-immune cell types, the latter including adipocytes, endothelial cells (EC), pericytes and myofibroblasts (MF). This matrix was used to perform CIBERSORT analysis (on-line access to the webserver http://cibersort.stanford.edu/) of GEP data from 3 publicly datasets (GSE10846, GSE19246 and GSE34171) of overall 604 DLBCL cases. The relative percentage of each tumor-infiltrating cell fraction was estimated and a global heatmap of tumor microenvironment composition was generated. Results were also stratified according to either cell of origin or clinical outcome data and significant differences in each subgroup calculated by a two-sided Mann-Whitney test.
Results
By applying CIBERSORT, we found that, among immune cells, memory B cells, plasma cells, NK cells, dendritic cells, CD4 T cells and macrophages mainly infiltrate DLBCL tissues. In particular, plasma cells, NK and dendritic cells showed significant higher proportion in ABC than GCB tumors, whereas CD4 T-cells and dendritic cells were prevalent in patients with better outcome. Notably, among stromal cells, MF was the most represented population and exhibited a significant predominance in patients with more favorable outcome and a strong positive correlation with overall survival (Figure 1). Consistently, the fraction of MF was significantly larger in GCB tumors compared with ABC subtypes. These latter resulted more enriched in EC.
Conclusion
By estimating relative fractions of DLBCL-infiltrating cells using CIBERSORT, we studied unknown aspects of tumor microenvironment with potential prognostic implications. Our data suggest that the composition of DLBCL with diverse molecular and clinical features also differ in term of immune and, particularly, stromal cell infiltration. These findings are in line with previous reports highlighting the prognostic value of specific stromal gene signatures and support further studies aimed at uncover novel prognostic/predictive biomarkers correlated with non-malignant tumor-associated cell subsets.

Session topic: E-poster
Keyword(s): Diffuse large B cell lymphoma, Microenvironment, Prognostic factor, Stromal cell
Type: Eposter Presentation
Background
Diffuse large B cell lymphoma (DLBCL) comprises a large group of disease entities with high molecular heterogeneity and variable treatment responsiveness. Results from gene expression profiling (GEP) studies highlighted the role of cell of origin, namely activated B cell-like cells (ABC) and germinal center B cells (GCB), and stromal gene signatures for predicting clinical outcome and stratifying patient risk. However, translation of GEP information in easily applicable prognostic biomarkers remains a challenge.
Aims
We applied an innovative GEP-based computational method, namely CIBERSORT, to determine the cellular composition of DLBCL microenvironment and investigate associations between abundance of certain tumor-infiltrating non-malignant cell types and molecular/clinical traits of the disease.
Methods
We generated a gene signature matrix including a total number of 1028 genes that distinguish 17 different immune and non-immune cell types, the latter including adipocytes, endothelial cells (EC), pericytes and myofibroblasts (MF). This matrix was used to perform CIBERSORT analysis (on-line access to the webserver http://cibersort.stanford.edu/) of GEP data from 3 publicly datasets (GSE10846, GSE19246 and GSE34171) of overall 604 DLBCL cases. The relative percentage of each tumor-infiltrating cell fraction was estimated and a global heatmap of tumor microenvironment composition was generated. Results were also stratified according to either cell of origin or clinical outcome data and significant differences in each subgroup calculated by a two-sided Mann-Whitney test.
Results
By applying CIBERSORT, we found that, among immune cells, memory B cells, plasma cells, NK cells, dendritic cells, CD4 T cells and macrophages mainly infiltrate DLBCL tissues. In particular, plasma cells, NK and dendritic cells showed significant higher proportion in ABC than GCB tumors, whereas CD4 T-cells and dendritic cells were prevalent in patients with better outcome. Notably, among stromal cells, MF was the most represented population and exhibited a significant predominance in patients with more favorable outcome and a strong positive correlation with overall survival (Figure 1). Consistently, the fraction of MF was significantly larger in GCB tumors compared with ABC subtypes. These latter resulted more enriched in EC.
Conclusion
By estimating relative fractions of DLBCL-infiltrating cells using CIBERSORT, we studied unknown aspects of tumor microenvironment with potential prognostic implications. Our data suggest that the composition of DLBCL with diverse molecular and clinical features also differ in term of immune and, particularly, stromal cell infiltration. These findings are in line with previous reports highlighting the prognostic value of specific stromal gene signatures and support further studies aimed at uncover novel prognostic/predictive biomarkers correlated with non-malignant tumor-associated cell subsets.

Session topic: E-poster
Keyword(s): Diffuse large B cell lymphoma, Microenvironment, Prognostic factor, Stromal cell
{{ help_message }}
{{filter}}


