MOLECULAR CHARACTERIZATION OF MYELOPROLIFERATIVE NEOPLASMS WITH CONCOMITANT BCR-ABL1 AND JAK2 V617F OR CALRETICULIN MUTATIONS
(Abstract release date: 05/19/16)
EHA Library. Bödör C. 06/09/16; 132880; E1331

Dr. Csaba Bödör
Contributions
Contributions
Abstract
Abstract: E1331
Type: Eposter Presentation
Background
Myeloproliferative neoplasms (MPN) are classified based on the presence of the BCR-ABL1, JAK2 and CALRETICULIN mutations. These are considered to be mutually exclusive, however rare cases with concomitant occurrence of these driver mutations have been reported, representing an ideal model to chronicle the molecular alterations and to investigate the clonal architecture in these cases. We report a comprehensive clinical and genetic analysis of sequential samples obtained from 7 patients with concurrent BCR-ABL1 and JAK2 V617F or CALR mutations diagnosed at our centers between 1998 and 2015.
Aims
To perform a detailed clinical, morphological and quantitative genetic analysis of sequential samples obtained from 7 patients with MPN with concomitant BCR-ABL1 and JAK2 V617F (n=5) or CALR mutations (n=2) to gain further insight into clonal architecture and pathogenesis of these rare double-mutated cases.
Methods
DNAs and RNAs extracted from peripheral blood or bone marrow samples were used to perform quantitative analysis of the BCR-ABL1 fusion transcript and JAK2 V617F/CALR mutations as part of the routine molecular diagnostic workup. The mutation status of 13 genes (JAK2, CALR, MPL, ASXL1, EZH2, SRSF2, IDH1, IDH2, LNK, CBL, TET2, DNMT3A, TP53) frequently mutated in MPNs was analyzed using next generation sequencing (NGS). The NGS analysis for performed using the AmpliSeq technology with an IonTorrent instrument at an average depth of 1000x on 15 sequential samples from 7 patients.
Results
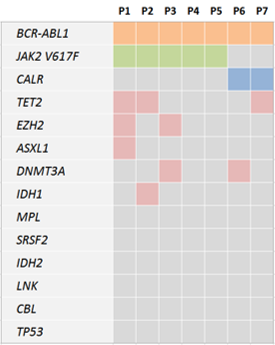
The quantitative analyses of the BCR-ABL1 fusion transcript and JAK2 V617F or CALR mutations suggested that these driver mutations occurred in different clones in 5 out the 7 cases analyzed. The next generation sequencing analysis of the 13 target genes identified additional mutations in 5 of the 7 cases with concurrent driver mutations (summarized in the heat map in Figure 1). The most frequently mutated gene was TET2 with mutations identified in 3 cases (2 cases with BCR-ABL1 and JAK2 mutation and one case with BCR-ABL1 and CALR mutation). Both EZH2 and DNMT3A mutations were detected 2 cases. ASXL1 and IDH1 were found to carry mutations in single cases with BCR-ABL1 and JAK2 mutation. The 5 patients harboring additional mutations carried 1-3 mutation per sample with one of the cases presenting with 3 additional mutations affecting the TET2, EZH2 and ASXL1 genes.
Conclusion
Our analysis of the 7 patients with concurrent MPN driver mutations suggested that the BCR-ABL1 and JAK2 or CALR mutations occur in different clones in majority of cases with frequent acquisition of additional phenotype modifying genes including TET2, EZH2, ASXL1 and DNMT3A.
Session topic: E-poster
Keyword(s): BCR-ABL, Myeloproliferative disorder
Type: Eposter Presentation
Background
Myeloproliferative neoplasms (MPN) are classified based on the presence of the BCR-ABL1, JAK2 and CALRETICULIN mutations. These are considered to be mutually exclusive, however rare cases with concomitant occurrence of these driver mutations have been reported, representing an ideal model to chronicle the molecular alterations and to investigate the clonal architecture in these cases. We report a comprehensive clinical and genetic analysis of sequential samples obtained from 7 patients with concurrent BCR-ABL1 and JAK2 V617F or CALR mutations diagnosed at our centers between 1998 and 2015.
Aims
To perform a detailed clinical, morphological and quantitative genetic analysis of sequential samples obtained from 7 patients with MPN with concomitant BCR-ABL1 and JAK2 V617F (n=5) or CALR mutations (n=2) to gain further insight into clonal architecture and pathogenesis of these rare double-mutated cases.
Methods
DNAs and RNAs extracted from peripheral blood or bone marrow samples were used to perform quantitative analysis of the BCR-ABL1 fusion transcript and JAK2 V617F/CALR mutations as part of the routine molecular diagnostic workup. The mutation status of 13 genes (JAK2, CALR, MPL, ASXL1, EZH2, SRSF2, IDH1, IDH2, LNK, CBL, TET2, DNMT3A, TP53) frequently mutated in MPNs was analyzed using next generation sequencing (NGS). The NGS analysis for performed using the AmpliSeq technology with an IonTorrent instrument at an average depth of 1000x on 15 sequential samples from 7 patients.
Results
The quantitative analyses of the BCR-ABL1 fusion transcript and JAK2 V617F or CALR mutations suggested that these driver mutations occurred in different clones in 5 out the 7 cases analyzed. The next generation sequencing analysis of the 13 target genes identified additional mutations in 5 of the 7 cases with concurrent driver mutations (summarized in the heat map in Figure 1). The most frequently mutated gene was TET2 with mutations identified in 3 cases (2 cases with BCR-ABL1 and JAK2 mutation and one case with BCR-ABL1 and CALR mutation). Both EZH2 and DNMT3A mutations were detected 2 cases. ASXL1 and IDH1 were found to carry mutations in single cases with BCR-ABL1 and JAK2 mutation. The 5 patients harboring additional mutations carried 1-3 mutation per sample with one of the cases presenting with 3 additional mutations affecting the TET2, EZH2 and ASXL1 genes.
Conclusion
Our analysis of the 7 patients with concurrent MPN driver mutations suggested that the BCR-ABL1 and JAK2 or CALR mutations occur in different clones in majority of cases with frequent acquisition of additional phenotype modifying genes including TET2, EZH2, ASXL1 and DNMT3A.

Session topic: E-poster
Keyword(s): BCR-ABL, Myeloproliferative disorder
Abstract: E1331
Type: Eposter Presentation
Background
Myeloproliferative neoplasms (MPN) are classified based on the presence of the BCR-ABL1, JAK2 and CALRETICULIN mutations. These are considered to be mutually exclusive, however rare cases with concomitant occurrence of these driver mutations have been reported, representing an ideal model to chronicle the molecular alterations and to investigate the clonal architecture in these cases. We report a comprehensive clinical and genetic analysis of sequential samples obtained from 7 patients with concurrent BCR-ABL1 and JAK2 V617F or CALR mutations diagnosed at our centers between 1998 and 2015.
Aims
To perform a detailed clinical, morphological and quantitative genetic analysis of sequential samples obtained from 7 patients with MPN with concomitant BCR-ABL1 and JAK2 V617F (n=5) or CALR mutations (n=2) to gain further insight into clonal architecture and pathogenesis of these rare double-mutated cases.
Methods
DNAs and RNAs extracted from peripheral blood or bone marrow samples were used to perform quantitative analysis of the BCR-ABL1 fusion transcript and JAK2 V617F/CALR mutations as part of the routine molecular diagnostic workup. The mutation status of 13 genes (JAK2, CALR, MPL, ASXL1, EZH2, SRSF2, IDH1, IDH2, LNK, CBL, TET2, DNMT3A, TP53) frequently mutated in MPNs was analyzed using next generation sequencing (NGS). The NGS analysis for performed using the AmpliSeq technology with an IonTorrent instrument at an average depth of 1000x on 15 sequential samples from 7 patients.
Results
The quantitative analyses of the BCR-ABL1 fusion transcript and JAK2 V617F or CALR mutations suggested that these driver mutations occurred in different clones in 5 out the 7 cases analyzed. The next generation sequencing analysis of the 13 target genes identified additional mutations in 5 of the 7 cases with concurrent driver mutations (summarized in the heat map in Figure 1). The most frequently mutated gene was TET2 with mutations identified in 3 cases (2 cases with BCR-ABL1 and JAK2 mutation and one case with BCR-ABL1 and CALR mutation). Both EZH2 and DNMT3A mutations were detected 2 cases. ASXL1 and IDH1 were found to carry mutations in single cases with BCR-ABL1 and JAK2 mutation. The 5 patients harboring additional mutations carried 1-3 mutation per sample with one of the cases presenting with 3 additional mutations affecting the TET2, EZH2 and ASXL1 genes.
Conclusion
Our analysis of the 7 patients with concurrent MPN driver mutations suggested that the BCR-ABL1 and JAK2 or CALR mutations occur in different clones in majority of cases with frequent acquisition of additional phenotype modifying genes including TET2, EZH2, ASXL1 and DNMT3A.

Session topic: E-poster
Keyword(s): BCR-ABL, Myeloproliferative disorder
Type: Eposter Presentation
Background
Myeloproliferative neoplasms (MPN) are classified based on the presence of the BCR-ABL1, JAK2 and CALRETICULIN mutations. These are considered to be mutually exclusive, however rare cases with concomitant occurrence of these driver mutations have been reported, representing an ideal model to chronicle the molecular alterations and to investigate the clonal architecture in these cases. We report a comprehensive clinical and genetic analysis of sequential samples obtained from 7 patients with concurrent BCR-ABL1 and JAK2 V617F or CALR mutations diagnosed at our centers between 1998 and 2015.
Aims
To perform a detailed clinical, morphological and quantitative genetic analysis of sequential samples obtained from 7 patients with MPN with concomitant BCR-ABL1 and JAK2 V617F (n=5) or CALR mutations (n=2) to gain further insight into clonal architecture and pathogenesis of these rare double-mutated cases.
Methods
DNAs and RNAs extracted from peripheral blood or bone marrow samples were used to perform quantitative analysis of the BCR-ABL1 fusion transcript and JAK2 V617F/CALR mutations as part of the routine molecular diagnostic workup. The mutation status of 13 genes (JAK2, CALR, MPL, ASXL1, EZH2, SRSF2, IDH1, IDH2, LNK, CBL, TET2, DNMT3A, TP53) frequently mutated in MPNs was analyzed using next generation sequencing (NGS). The NGS analysis for performed using the AmpliSeq technology with an IonTorrent instrument at an average depth of 1000x on 15 sequential samples from 7 patients.
Results
The quantitative analyses of the BCR-ABL1 fusion transcript and JAK2 V617F or CALR mutations suggested that these driver mutations occurred in different clones in 5 out the 7 cases analyzed. The next generation sequencing analysis of the 13 target genes identified additional mutations in 5 of the 7 cases with concurrent driver mutations (summarized in the heat map in Figure 1). The most frequently mutated gene was TET2 with mutations identified in 3 cases (2 cases with BCR-ABL1 and JAK2 mutation and one case with BCR-ABL1 and CALR mutation). Both EZH2 and DNMT3A mutations were detected 2 cases. ASXL1 and IDH1 were found to carry mutations in single cases with BCR-ABL1 and JAK2 mutation. The 5 patients harboring additional mutations carried 1-3 mutation per sample with one of the cases presenting with 3 additional mutations affecting the TET2, EZH2 and ASXL1 genes.
Conclusion
Our analysis of the 7 patients with concurrent MPN driver mutations suggested that the BCR-ABL1 and JAK2 or CALR mutations occur in different clones in majority of cases with frequent acquisition of additional phenotype modifying genes including TET2, EZH2, ASXL1 and DNMT3A.

Session topic: E-poster
Keyword(s): BCR-ABL, Myeloproliferative disorder
{{ help_message }}
{{filter}}


