GENETIC FACTORS ASSOCIATED WITH EVOLUTION OF MYELODYSPLASTIC SYNDROMES TO SECONDARY CHRONIC MYELOMONOCYTIC LEUKEMIA.
(Abstract release date: 05/19/16)
EHA Library. Saiki R. 06/09/16; 132745; E1196

Mr. Ryunosuke Saiki
Contributions
Contributions
Abstract
Abstract: E1196
Type: Eposter Presentation
Background
Chronic myelomonocytic leukemia (CMML) is a subset of myelodysplastic/myeloproliferative neoplasms (MDS/MPN), which are morphologically distinct from myelodysplastic syndromes (MDS). Nevertheless, CMML and MDS share several genetic defects and dysplastic features. Moreover, CMML can evolve from MDS, designated as secondary CMML (sCMML). The molecular characteristics of sCMML, which are distinct from those of primary CMML, remain to be elucidated.
Aims
By comparing the genetic/cytogenetic abnormalities between sCMML and other myeloid malignancies, we clarify the molecular pathogenesis of sCMML.
Methods
A total of 601 cases with different myeloid neoplasms, including de novo CMML (n=89), sCMML (n=26), MDS (n=348), and secondary AML (sAML) (n=138) were subjected to the analysis, combining the genotyping data from our cohort and publicly available datasets. Serial samples were available for 7 cases.
Results
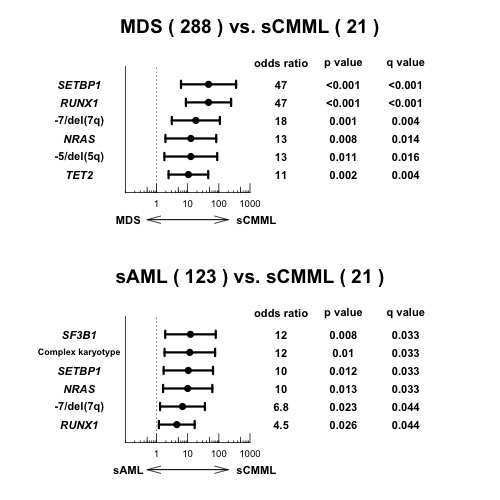
In the de novo CMML cohort, the most frequently mutated genes included TET2 (40%), SRSF2 (30%), ASXL1 (24%), RUNX1 (16%), CBL (11%), EZH2 (11%), ZRSR2 (11%), and KRAS (10%), which, combined, accounted for 74% of the CMML cases. In the sCMML cohort, by contrast, RUNX1 (32%), TET2 (32%), ASXL1 (24%), SF3B1 (24%), and SETBP1 (20%) represented common genetic alterations, together with -7/del(7q) (43%), complex karyotype (38%), and del(5q) (24%). In multivariate analysis comparing the spectrum of mutations between sCMML and de novo CMML, SF3B1, and RUNX1 mutations were enriched in sCMML patients. In patients with sCMML, SF3B1 mutations supposed to be acquired prior to RUNX1 mutations based on the comparison of their variant allele frequencies. Most likely, SF3B1 mutations were acquired as founder events in the MDS phase and RUNX1 mutations occurred during sCMML evolution. However, RUNX1 mutations were not always identified in patients with sCMML, suggesting that other causes are also related to sCMML evolution.To further identify genetic lesions specifically associated with CMML evolution, we compared the spectrum of mutations between sCMML and MDS. Multivariate analysis revealed that mutations in SETBP1, RUNX1, TET2, and NRAS, del(5q), and -7/del(7q) were significantly and independently associated with sCMML. Then we divided de novo CMML cases into 2 categories; Type-1 with ≥2 of these lesions (n=13) and Type-2 with <2 (n=54). Patients with Type-1 CMML had a significantly shorter overall survival compared to Type-2 disease (HR=3.73, 95%CI 1.78-7.79, P<0.001). Serial sample analysis revealed that mutations in SETBP1, RUNX1, and NRAS, and del(7q) tended to be newly acquired during evolution to sCMML.However, the lesions extracted in the comparison between sCMML and MDS were also detected in our cohort of sAML; -7/del(7q) (23%), del(5q) (20%), mutations in SETBP1 (4%), RUNX1 (19%), TET2 (22%), and NRAS (7%). To investigate what lesions are really characteristic to sCMML evolution from MDS, we compared the frequencies of genetic lesions between sCMML and sAML. We found SF3B1, SETBP1, NRAS, and RUNX1 mutations, as well as complex karyotype, and -7/del(7q) were significantly enriched in sCMML, which largely overlapped with the lesions associated with evolution from MDS to sCMML.
Conclusion
In this study, a novel set of lesions associated with sCMML evolution (SETBP1, RUNX1, TET2, and NRAS mutations, del(5q), and -7/del(7q)) was identified and shown to be able to stratify de novo CMML into clinically distinct subgroups. By genotyping serial samples and comparing genetic lesions between sCMML and sAML, this set of lesions was thought to be characteristic to sCMML evolution. Our study demonstrated that specific genetic events might play an important role in sCMML evolution from MDS.
Session topic: E-poster
Keyword(s): Chronic myelomonocytic leukemia, MDS, Mutation status, Secondary
Type: Eposter Presentation
Background
Chronic myelomonocytic leukemia (CMML) is a subset of myelodysplastic/myeloproliferative neoplasms (MDS/MPN), which are morphologically distinct from myelodysplastic syndromes (MDS). Nevertheless, CMML and MDS share several genetic defects and dysplastic features. Moreover, CMML can evolve from MDS, designated as secondary CMML (sCMML). The molecular characteristics of sCMML, which are distinct from those of primary CMML, remain to be elucidated.
Aims
By comparing the genetic/cytogenetic abnormalities between sCMML and other myeloid malignancies, we clarify the molecular pathogenesis of sCMML.
Methods
A total of 601 cases with different myeloid neoplasms, including de novo CMML (n=89), sCMML (n=26), MDS (n=348), and secondary AML (sAML) (n=138) were subjected to the analysis, combining the genotyping data from our cohort and publicly available datasets. Serial samples were available for 7 cases.
Results
In the de novo CMML cohort, the most frequently mutated genes included TET2 (40%), SRSF2 (30%), ASXL1 (24%), RUNX1 (16%), CBL (11%), EZH2 (11%), ZRSR2 (11%), and KRAS (10%), which, combined, accounted for 74% of the CMML cases. In the sCMML cohort, by contrast, RUNX1 (32%), TET2 (32%), ASXL1 (24%), SF3B1 (24%), and SETBP1 (20%) represented common genetic alterations, together with -7/del(7q) (43%), complex karyotype (38%), and del(5q) (24%). In multivariate analysis comparing the spectrum of mutations between sCMML and de novo CMML, SF3B1, and RUNX1 mutations were enriched in sCMML patients. In patients with sCMML, SF3B1 mutations supposed to be acquired prior to RUNX1 mutations based on the comparison of their variant allele frequencies. Most likely, SF3B1 mutations were acquired as founder events in the MDS phase and RUNX1 mutations occurred during sCMML evolution. However, RUNX1 mutations were not always identified in patients with sCMML, suggesting that other causes are also related to sCMML evolution.To further identify genetic lesions specifically associated with CMML evolution, we compared the spectrum of mutations between sCMML and MDS. Multivariate analysis revealed that mutations in SETBP1, RUNX1, TET2, and NRAS, del(5q), and -7/del(7q) were significantly and independently associated with sCMML. Then we divided de novo CMML cases into 2 categories; Type-1 with ≥2 of these lesions (n=13) and Type-2 with <2 (n=54). Patients with Type-1 CMML had a significantly shorter overall survival compared to Type-2 disease (HR=3.73, 95%CI 1.78-7.79, P<0.001). Serial sample analysis revealed that mutations in SETBP1, RUNX1, and NRAS, and del(7q) tended to be newly acquired during evolution to sCMML.However, the lesions extracted in the comparison between sCMML and MDS were also detected in our cohort of sAML; -7/del(7q) (23%), del(5q) (20%), mutations in SETBP1 (4%), RUNX1 (19%), TET2 (22%), and NRAS (7%). To investigate what lesions are really characteristic to sCMML evolution from MDS, we compared the frequencies of genetic lesions between sCMML and sAML. We found SF3B1, SETBP1, NRAS, and RUNX1 mutations, as well as complex karyotype, and -7/del(7q) were significantly enriched in sCMML, which largely overlapped with the lesions associated with evolution from MDS to sCMML.
Conclusion
In this study, a novel set of lesions associated with sCMML evolution (SETBP1, RUNX1, TET2, and NRAS mutations, del(5q), and -7/del(7q)) was identified and shown to be able to stratify de novo CMML into clinically distinct subgroups. By genotyping serial samples and comparing genetic lesions between sCMML and sAML, this set of lesions was thought to be characteristic to sCMML evolution. Our study demonstrated that specific genetic events might play an important role in sCMML evolution from MDS.

Session topic: E-poster
Keyword(s): Chronic myelomonocytic leukemia, MDS, Mutation status, Secondary
Abstract: E1196
Type: Eposter Presentation
Background
Chronic myelomonocytic leukemia (CMML) is a subset of myelodysplastic/myeloproliferative neoplasms (MDS/MPN), which are morphologically distinct from myelodysplastic syndromes (MDS). Nevertheless, CMML and MDS share several genetic defects and dysplastic features. Moreover, CMML can evolve from MDS, designated as secondary CMML (sCMML). The molecular characteristics of sCMML, which are distinct from those of primary CMML, remain to be elucidated.
Aims
By comparing the genetic/cytogenetic abnormalities between sCMML and other myeloid malignancies, we clarify the molecular pathogenesis of sCMML.
Methods
A total of 601 cases with different myeloid neoplasms, including de novo CMML (n=89), sCMML (n=26), MDS (n=348), and secondary AML (sAML) (n=138) were subjected to the analysis, combining the genotyping data from our cohort and publicly available datasets. Serial samples were available for 7 cases.
Results
In the de novo CMML cohort, the most frequently mutated genes included TET2 (40%), SRSF2 (30%), ASXL1 (24%), RUNX1 (16%), CBL (11%), EZH2 (11%), ZRSR2 (11%), and KRAS (10%), which, combined, accounted for 74% of the CMML cases. In the sCMML cohort, by contrast, RUNX1 (32%), TET2 (32%), ASXL1 (24%), SF3B1 (24%), and SETBP1 (20%) represented common genetic alterations, together with -7/del(7q) (43%), complex karyotype (38%), and del(5q) (24%). In multivariate analysis comparing the spectrum of mutations between sCMML and de novo CMML, SF3B1, and RUNX1 mutations were enriched in sCMML patients. In patients with sCMML, SF3B1 mutations supposed to be acquired prior to RUNX1 mutations based on the comparison of their variant allele frequencies. Most likely, SF3B1 mutations were acquired as founder events in the MDS phase and RUNX1 mutations occurred during sCMML evolution. However, RUNX1 mutations were not always identified in patients with sCMML, suggesting that other causes are also related to sCMML evolution.To further identify genetic lesions specifically associated with CMML evolution, we compared the spectrum of mutations between sCMML and MDS. Multivariate analysis revealed that mutations in SETBP1, RUNX1, TET2, and NRAS, del(5q), and -7/del(7q) were significantly and independently associated with sCMML. Then we divided de novo CMML cases into 2 categories; Type-1 with ≥2 of these lesions (n=13) and Type-2 with <2 (n=54). Patients with Type-1 CMML had a significantly shorter overall survival compared to Type-2 disease (HR=3.73, 95%CI 1.78-7.79, P<0.001). Serial sample analysis revealed that mutations in SETBP1, RUNX1, and NRAS, and del(7q) tended to be newly acquired during evolution to sCMML.However, the lesions extracted in the comparison between sCMML and MDS were also detected in our cohort of sAML; -7/del(7q) (23%), del(5q) (20%), mutations in SETBP1 (4%), RUNX1 (19%), TET2 (22%), and NRAS (7%). To investigate what lesions are really characteristic to sCMML evolution from MDS, we compared the frequencies of genetic lesions between sCMML and sAML. We found SF3B1, SETBP1, NRAS, and RUNX1 mutations, as well as complex karyotype, and -7/del(7q) were significantly enriched in sCMML, which largely overlapped with the lesions associated with evolution from MDS to sCMML.
Conclusion
In this study, a novel set of lesions associated with sCMML evolution (SETBP1, RUNX1, TET2, and NRAS mutations, del(5q), and -7/del(7q)) was identified and shown to be able to stratify de novo CMML into clinically distinct subgroups. By genotyping serial samples and comparing genetic lesions between sCMML and sAML, this set of lesions was thought to be characteristic to sCMML evolution. Our study demonstrated that specific genetic events might play an important role in sCMML evolution from MDS.

Session topic: E-poster
Keyword(s): Chronic myelomonocytic leukemia, MDS, Mutation status, Secondary
Type: Eposter Presentation
Background
Chronic myelomonocytic leukemia (CMML) is a subset of myelodysplastic/myeloproliferative neoplasms (MDS/MPN), which are morphologically distinct from myelodysplastic syndromes (MDS). Nevertheless, CMML and MDS share several genetic defects and dysplastic features. Moreover, CMML can evolve from MDS, designated as secondary CMML (sCMML). The molecular characteristics of sCMML, which are distinct from those of primary CMML, remain to be elucidated.
Aims
By comparing the genetic/cytogenetic abnormalities between sCMML and other myeloid malignancies, we clarify the molecular pathogenesis of sCMML.
Methods
A total of 601 cases with different myeloid neoplasms, including de novo CMML (n=89), sCMML (n=26), MDS (n=348), and secondary AML (sAML) (n=138) were subjected to the analysis, combining the genotyping data from our cohort and publicly available datasets. Serial samples were available for 7 cases.
Results
In the de novo CMML cohort, the most frequently mutated genes included TET2 (40%), SRSF2 (30%), ASXL1 (24%), RUNX1 (16%), CBL (11%), EZH2 (11%), ZRSR2 (11%), and KRAS (10%), which, combined, accounted for 74% of the CMML cases. In the sCMML cohort, by contrast, RUNX1 (32%), TET2 (32%), ASXL1 (24%), SF3B1 (24%), and SETBP1 (20%) represented common genetic alterations, together with -7/del(7q) (43%), complex karyotype (38%), and del(5q) (24%). In multivariate analysis comparing the spectrum of mutations between sCMML and de novo CMML, SF3B1, and RUNX1 mutations were enriched in sCMML patients. In patients with sCMML, SF3B1 mutations supposed to be acquired prior to RUNX1 mutations based on the comparison of their variant allele frequencies. Most likely, SF3B1 mutations were acquired as founder events in the MDS phase and RUNX1 mutations occurred during sCMML evolution. However, RUNX1 mutations were not always identified in patients with sCMML, suggesting that other causes are also related to sCMML evolution.To further identify genetic lesions specifically associated with CMML evolution, we compared the spectrum of mutations between sCMML and MDS. Multivariate analysis revealed that mutations in SETBP1, RUNX1, TET2, and NRAS, del(5q), and -7/del(7q) were significantly and independently associated with sCMML. Then we divided de novo CMML cases into 2 categories; Type-1 with ≥2 of these lesions (n=13) and Type-2 with <2 (n=54). Patients with Type-1 CMML had a significantly shorter overall survival compared to Type-2 disease (HR=3.73, 95%CI 1.78-7.79, P<0.001). Serial sample analysis revealed that mutations in SETBP1, RUNX1, and NRAS, and del(7q) tended to be newly acquired during evolution to sCMML.However, the lesions extracted in the comparison between sCMML and MDS were also detected in our cohort of sAML; -7/del(7q) (23%), del(5q) (20%), mutations in SETBP1 (4%), RUNX1 (19%), TET2 (22%), and NRAS (7%). To investigate what lesions are really characteristic to sCMML evolution from MDS, we compared the frequencies of genetic lesions between sCMML and sAML. We found SF3B1, SETBP1, NRAS, and RUNX1 mutations, as well as complex karyotype, and -7/del(7q) were significantly enriched in sCMML, which largely overlapped with the lesions associated with evolution from MDS to sCMML.
Conclusion
In this study, a novel set of lesions associated with sCMML evolution (SETBP1, RUNX1, TET2, and NRAS mutations, del(5q), and -7/del(7q)) was identified and shown to be able to stratify de novo CMML into clinically distinct subgroups. By genotyping serial samples and comparing genetic lesions between sCMML and sAML, this set of lesions was thought to be characteristic to sCMML evolution. Our study demonstrated that specific genetic events might play an important role in sCMML evolution from MDS.

Session topic: E-poster
Keyword(s): Chronic myelomonocytic leukemia, MDS, Mutation status, Secondary
{{ help_message }}
{{filter}}


