SINGLE NUCLEOTIDE POLYMORPHISMS IN DRUG-RELATED GENES ARE ASSOCIATED WITH RESISTANCE OR TOXICITY TO IMATINIB IN CHRONIC MYELOID LEUKAEMIA
(Abstract release date: 05/19/16)
EHA Library. Estrada N. 06/09/16; 132641; E1092
Disclosure(s): Authors declare no conflicts of interest

Mrs. Natalia Estrada
Contributions
Contributions
Abstract
Abstract: E1092
Type: Eposter Presentation
Background
Imatinib (IM) induces apoptosis in BCR-ABL1 positive cell lines and is extensively used to treat patients with newly diagnosed Philadelphia chromosome-positive Chronic Myeloid Leukaemia (Ph+CML). However, about 20% of Ph+CML patients fail to achieve optimal response, and a substantial proportion develop intolerance or toxicity. Amplification and overexpression of the BCR-ABL1 gene, BCR-ABL1 kinase domain point mutations and genetic variability (as genetic polymorphisms), among others, may explain IM resistance.
Aims
To assess the association between single-nucleotide polymorphisms (SNP) in drug-related genes and IM resistance or toxicity in CML patients.
Methods
In a pilot study with 45 Ph+CML patients treated with IM from three Spanish centers, we tested 1936 SNPs in 225 drug-related genes using the Affymetrix Drug Metabolism Enzymes and Transporters (DMET) microarray. Response criteria were assessed according to ELN2009 or ELN2013 guidelines. Relevant toxicity to IM was defined as the need for switching to another tyrosine kinase inhibitor (TKI).SNPs significantly associated with response or toxicity to IM treatment were validated in an independent group of 105 Ph+CML patients from 8 Spanish centers. Informed consent (according to the Declaration of Helsinki) was obtained from all patients. Validation genotyping was performed using Fluidigm 96.96 Dynamic Array with BioMark HD Systems (Fluidigm Corp., CA). Data were analysed by the BioMark SNP Genotyping Analysis software to obtain genotype calls. Candidate SNPs were primarily evaluated for adequacy of Hardy-Weinber Equilibrium (HWE) using the Chi-square test. For the association study, the SNPassoc package from R was used (PMID: 17267436, www.r-project.com).
Results
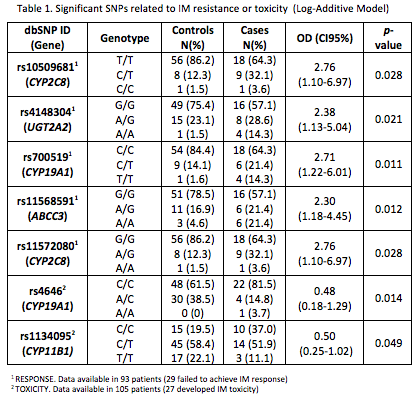
In this pilot study we found 76 SNPs to be significantly associated with response or toxicity to IM treatment. In the validation cohort, 5/76 SNPs and 2/76 SNPs were related to IM resistance or toxicity, respectively (Table 1). Patients carrying at least one variant allele for CYP2C8 (C), UGT2A2 (A), CYP19A1 (T), ABCC3 (A) and CYP2C8 (A) were significantly associated with IM treatment failure. In the same way, AC-AA genotypes in CYP19A1 and CT-TT genotypes in CYP11B1 were significantly associated with lower risk of IM-toxicity.
Conclusion
The SNPs found in the present study allow the identification of CML patients at risk to develop IM resistance or toxicity, suggesting that a second generation TKI should be offered to these patients as first line therapy. Furthermore, our results suggest that SNP analysis may be included in clinical practice as a predictive tool for response and toxicity at diagnosis of CML patients. Acknowledgements: This study was supported by an unrestricted grant from Novartis.
Session topic: E-poster
Keyword(s): Chronic myeloid leukemia, Imatinib, Microarray analysis, Single nucleotide polymorphism
Type: Eposter Presentation
Background
Imatinib (IM) induces apoptosis in BCR-ABL1 positive cell lines and is extensively used to treat patients with newly diagnosed Philadelphia chromosome-positive Chronic Myeloid Leukaemia (Ph+CML). However, about 20% of Ph+CML patients fail to achieve optimal response, and a substantial proportion develop intolerance or toxicity. Amplification and overexpression of the BCR-ABL1 gene, BCR-ABL1 kinase domain point mutations and genetic variability (as genetic polymorphisms), among others, may explain IM resistance.
Aims
To assess the association between single-nucleotide polymorphisms (SNP) in drug-related genes and IM resistance or toxicity in CML patients.
Methods
In a pilot study with 45 Ph+CML patients treated with IM from three Spanish centers, we tested 1936 SNPs in 225 drug-related genes using the Affymetrix Drug Metabolism Enzymes and Transporters (DMET) microarray. Response criteria were assessed according to ELN2009 or ELN2013 guidelines. Relevant toxicity to IM was defined as the need for switching to another tyrosine kinase inhibitor (TKI).SNPs significantly associated with response or toxicity to IM treatment were validated in an independent group of 105 Ph+CML patients from 8 Spanish centers. Informed consent (according to the Declaration of Helsinki) was obtained from all patients. Validation genotyping was performed using Fluidigm 96.96 Dynamic Array with BioMark HD Systems (Fluidigm Corp., CA). Data were analysed by the BioMark SNP Genotyping Analysis software to obtain genotype calls. Candidate SNPs were primarily evaluated for adequacy of Hardy-Weinber Equilibrium (HWE) using the Chi-square test. For the association study, the SNPassoc package from R was used (PMID: 17267436, www.r-project.com).
Results
In this pilot study we found 76 SNPs to be significantly associated with response or toxicity to IM treatment. In the validation cohort, 5/76 SNPs and 2/76 SNPs were related to IM resistance or toxicity, respectively (Table 1). Patients carrying at least one variant allele for CYP2C8 (C), UGT2A2 (A), CYP19A1 (T), ABCC3 (A) and CYP2C8 (A) were significantly associated with IM treatment failure. In the same way, AC-AA genotypes in CYP19A1 and CT-TT genotypes in CYP11B1 were significantly associated with lower risk of IM-toxicity.
Conclusion
The SNPs found in the present study allow the identification of CML patients at risk to develop IM resistance or toxicity, suggesting that a second generation TKI should be offered to these patients as first line therapy. Furthermore, our results suggest that SNP analysis may be included in clinical practice as a predictive tool for response and toxicity at diagnosis of CML patients. Acknowledgements: This study was supported by an unrestricted grant from Novartis.

Session topic: E-poster
Keyword(s): Chronic myeloid leukemia, Imatinib, Microarray analysis, Single nucleotide polymorphism
Abstract: E1092
Type: Eposter Presentation
Background
Imatinib (IM) induces apoptosis in BCR-ABL1 positive cell lines and is extensively used to treat patients with newly diagnosed Philadelphia chromosome-positive Chronic Myeloid Leukaemia (Ph+CML). However, about 20% of Ph+CML patients fail to achieve optimal response, and a substantial proportion develop intolerance or toxicity. Amplification and overexpression of the BCR-ABL1 gene, BCR-ABL1 kinase domain point mutations and genetic variability (as genetic polymorphisms), among others, may explain IM resistance.
Aims
To assess the association between single-nucleotide polymorphisms (SNP) in drug-related genes and IM resistance or toxicity in CML patients.
Methods
In a pilot study with 45 Ph+CML patients treated with IM from three Spanish centers, we tested 1936 SNPs in 225 drug-related genes using the Affymetrix Drug Metabolism Enzymes and Transporters (DMET) microarray. Response criteria were assessed according to ELN2009 or ELN2013 guidelines. Relevant toxicity to IM was defined as the need for switching to another tyrosine kinase inhibitor (TKI).SNPs significantly associated with response or toxicity to IM treatment were validated in an independent group of 105 Ph+CML patients from 8 Spanish centers. Informed consent (according to the Declaration of Helsinki) was obtained from all patients. Validation genotyping was performed using Fluidigm 96.96 Dynamic Array with BioMark HD Systems (Fluidigm Corp., CA). Data were analysed by the BioMark SNP Genotyping Analysis software to obtain genotype calls. Candidate SNPs were primarily evaluated for adequacy of Hardy-Weinber Equilibrium (HWE) using the Chi-square test. For the association study, the SNPassoc package from R was used (PMID: 17267436, www.r-project.com).
Results
In this pilot study we found 76 SNPs to be significantly associated with response or toxicity to IM treatment. In the validation cohort, 5/76 SNPs and 2/76 SNPs were related to IM resistance or toxicity, respectively (Table 1). Patients carrying at least one variant allele for CYP2C8 (C), UGT2A2 (A), CYP19A1 (T), ABCC3 (A) and CYP2C8 (A) were significantly associated with IM treatment failure. In the same way, AC-AA genotypes in CYP19A1 and CT-TT genotypes in CYP11B1 were significantly associated with lower risk of IM-toxicity.
Conclusion
The SNPs found in the present study allow the identification of CML patients at risk to develop IM resistance or toxicity, suggesting that a second generation TKI should be offered to these patients as first line therapy. Furthermore, our results suggest that SNP analysis may be included in clinical practice as a predictive tool for response and toxicity at diagnosis of CML patients. Acknowledgements: This study was supported by an unrestricted grant from Novartis.

Session topic: E-poster
Keyword(s): Chronic myeloid leukemia, Imatinib, Microarray analysis, Single nucleotide polymorphism
Type: Eposter Presentation
Background
Imatinib (IM) induces apoptosis in BCR-ABL1 positive cell lines and is extensively used to treat patients with newly diagnosed Philadelphia chromosome-positive Chronic Myeloid Leukaemia (Ph+CML). However, about 20% of Ph+CML patients fail to achieve optimal response, and a substantial proportion develop intolerance or toxicity. Amplification and overexpression of the BCR-ABL1 gene, BCR-ABL1 kinase domain point mutations and genetic variability (as genetic polymorphisms), among others, may explain IM resistance.
Aims
To assess the association between single-nucleotide polymorphisms (SNP) in drug-related genes and IM resistance or toxicity in CML patients.
Methods
In a pilot study with 45 Ph+CML patients treated with IM from three Spanish centers, we tested 1936 SNPs in 225 drug-related genes using the Affymetrix Drug Metabolism Enzymes and Transporters (DMET) microarray. Response criteria were assessed according to ELN2009 or ELN2013 guidelines. Relevant toxicity to IM was defined as the need for switching to another tyrosine kinase inhibitor (TKI).SNPs significantly associated with response or toxicity to IM treatment were validated in an independent group of 105 Ph+CML patients from 8 Spanish centers. Informed consent (according to the Declaration of Helsinki) was obtained from all patients. Validation genotyping was performed using Fluidigm 96.96 Dynamic Array with BioMark HD Systems (Fluidigm Corp., CA). Data were analysed by the BioMark SNP Genotyping Analysis software to obtain genotype calls. Candidate SNPs were primarily evaluated for adequacy of Hardy-Weinber Equilibrium (HWE) using the Chi-square test. For the association study, the SNPassoc package from R was used (PMID: 17267436, www.r-project.com).
Results
In this pilot study we found 76 SNPs to be significantly associated with response or toxicity to IM treatment. In the validation cohort, 5/76 SNPs and 2/76 SNPs were related to IM resistance or toxicity, respectively (Table 1). Patients carrying at least one variant allele for CYP2C8 (C), UGT2A2 (A), CYP19A1 (T), ABCC3 (A) and CYP2C8 (A) were significantly associated with IM treatment failure. In the same way, AC-AA genotypes in CYP19A1 and CT-TT genotypes in CYP11B1 were significantly associated with lower risk of IM-toxicity.
Conclusion
The SNPs found in the present study allow the identification of CML patients at risk to develop IM resistance or toxicity, suggesting that a second generation TKI should be offered to these patients as first line therapy. Furthermore, our results suggest that SNP analysis may be included in clinical practice as a predictive tool for response and toxicity at diagnosis of CML patients. Acknowledgements: This study was supported by an unrestricted grant from Novartis.

Session topic: E-poster
Keyword(s): Chronic myeloid leukemia, Imatinib, Microarray analysis, Single nucleotide polymorphism
{{ help_message }}
{{filter}}


