DROPLED DIGITAL PCR MAY HAVE A PROGNOSTIC VALUE FOR PREDICTING RELAPSE AFTER IMATINIB DISCONTINUATION
(Abstract release date: 05/19/16)
EHA Library. Fava C. 06/09/16; 132639; E1090

Dr. Carmen Fava
Contributions
Contributions
Abstract
Abstract: E1090
Type: Eposter Presentation
Background
Treatment-free remission is a major goal in CML. Nowadays it is possible to safely discontinue imatinib but it is still not clear which patient (pt) will relapse. High sensitivity techniques like droplet digital (dd)PCR may help to discriminate pts who still present a significant amount of disease despite being in MR4 by standard real-time quantitative PCR (RQ-PCR).
Aims
To evaluate the capability of ddPCR to predict relapse after imatinib discontinuation in CML pts with stable MR4 by RQ-PCR.
Methods
Total RNA was extracted at different time-points from 19 pts in stable MR4 for two years before discontinuation of imatinib. ddPCR was carried out in triplicate using 200 ng of cDNA for each replicates and droplets were analyzed by QX100™ droplet reader (Bio-Rad) using the DigiDrop P210 Master Mix Kit (Bioclarma). All results were expressed according to the new recommendations [Cross, Leukemia 2015].The Wilcoxon test was used for the comparison of the total ABL medians; the Agreement Coefficient of the 2 methods was calculated with Cohen’s K test; Logistic Regression was used to assess the relationship between the probability of relapse and disease level by RQ-PCR and ddPCR.
Results
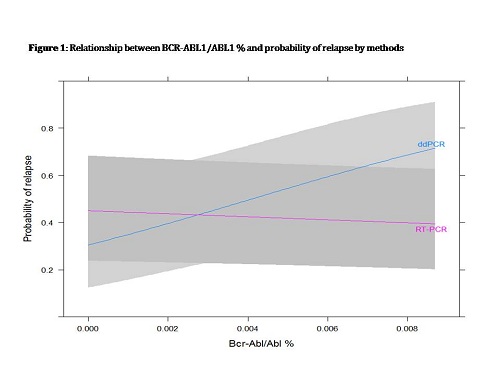
A total of 48 samples were retrospectively analyzed by RQ-PCR and ddPCR. All pts discontinued imatinib; 8/19 pts had to restart treatment for loss of MMR/MR4 after a median of 5.3 mos (2.2-8.5). A difference between ddPCR and RQ-PCR median total ABL copies (p<0.001) was found with the following medians (ranges): 46290 (8420-124600) and 93060 (25440-210000) for RQ-PCR and ddPCR respectively. A difference between ddPCR and RQ-PCR median total BCR-ABL1/ABL1% (p<0.001) was found: 0 (0-0.09); and 0.0009 (0-0.015) for RQ-PCR and ddPCR respectively. Sixteen pts had >2 evaluations, 3 had an evaluation between 14 and 18.5 mos before discontinuation. 6 pts had an evaluation at -12 mos (+/-0.81), 10 at -9 mos(+/-1.3), 14 at -6 mos(+/-0.9), 15 at -3 mos(+/-1.2), considering as time “0” the date of discontinuation. At time -12, only 1pt resulted BCR-ABL1 positive by both methods, 1 by ddPCR only and 0 by RQ-PCR only (K0.6, moderate agreement, p 0.13). At time -9, 3 pts resulted BCR-ABL1 positive by both methods, 6 by ddPCR only and none by RQ-PCR only (K=0.1, slight agreement, p 0.34). At time -6, 3 pts resulted BCR-ABL1 positive by both methods, 5 pts by ddPCR only and none by RQ-PCR only, (K=0.3, fair agreement, p 0.08). At time -3, none resulted BCR-ABL1 positive by both methods, 4 pts were positive by ddPCR only and 1 by RQ-PCR only (K=-0.1, no agreement, p 0.62). Over one year of molecular follow-up before discontinuation, for negative values obtained measuring BCR-ABL1 by RQ-PCR the probability of relapse was equal to about 50% (45%, Figure 1), i.e. the capability of RQ-PCR of detecting relapse was basically same as random being almost constant for all the positive values of BCR-ABL1. On the contrary the probability of a relapse for negative values of BCR-ABL1 by ddPCR was about 30% and it grew up linearly at increasing level of transcript, achieving a probability of 69% when BCR-ABL1/ABL1% was equal to 0.008%. The difference between RQ-PCR and ddPCR in predicting the probability of relapse was statistically significant (p 0.02).
Conclusion
Despite the small number of cases, our results confirm that the ddPCR is more sensitive than RQ-PCR for low levels of disease. Molecular follow-up by ddPCR at several time-points before discontinuation may have a prognostic value for predicting relapse, and this is worthy to be tested prospectively and in larger cohorts of pts.
Session topic: E-poster
Keyword(s): BCR-ABL, Molecular remission, MRD, PCR
Type: Eposter Presentation
Background
Treatment-free remission is a major goal in CML. Nowadays it is possible to safely discontinue imatinib but it is still not clear which patient (pt) will relapse. High sensitivity techniques like droplet digital (dd)PCR may help to discriminate pts who still present a significant amount of disease despite being in MR4 by standard real-time quantitative PCR (RQ-PCR).
Aims
To evaluate the capability of ddPCR to predict relapse after imatinib discontinuation in CML pts with stable MR4 by RQ-PCR.
Methods
Total RNA was extracted at different time-points from 19 pts in stable MR4 for two years before discontinuation of imatinib. ddPCR was carried out in triplicate using 200 ng of cDNA for each replicates and droplets were analyzed by QX100™ droplet reader (Bio-Rad) using the DigiDrop P210 Master Mix Kit (Bioclarma). All results were expressed according to the new recommendations [Cross, Leukemia 2015].The Wilcoxon test was used for the comparison of the total ABL medians; the Agreement Coefficient of the 2 methods was calculated with Cohen’s K test; Logistic Regression was used to assess the relationship between the probability of relapse and disease level by RQ-PCR and ddPCR.
Results
A total of 48 samples were retrospectively analyzed by RQ-PCR and ddPCR. All pts discontinued imatinib; 8/19 pts had to restart treatment for loss of MMR/MR4 after a median of 5.3 mos (2.2-8.5). A difference between ddPCR and RQ-PCR median total ABL copies (p<0.001) was found with the following medians (ranges): 46290 (8420-124600) and 93060 (25440-210000) for RQ-PCR and ddPCR respectively. A difference between ddPCR and RQ-PCR median total BCR-ABL1/ABL1% (p<0.001) was found: 0 (0-0.09); and 0.0009 (0-0.015) for RQ-PCR and ddPCR respectively. Sixteen pts had >2 evaluations, 3 had an evaluation between 14 and 18.5 mos before discontinuation. 6 pts had an evaluation at -12 mos (+/-0.81), 10 at -9 mos(+/-1.3), 14 at -6 mos(+/-0.9), 15 at -3 mos(+/-1.2), considering as time “0” the date of discontinuation. At time -12, only 1pt resulted BCR-ABL1 positive by both methods, 1 by ddPCR only and 0 by RQ-PCR only (K0.6, moderate agreement, p 0.13). At time -9, 3 pts resulted BCR-ABL1 positive by both methods, 6 by ddPCR only and none by RQ-PCR only (K=0.1, slight agreement, p 0.34). At time -6, 3 pts resulted BCR-ABL1 positive by both methods, 5 pts by ddPCR only and none by RQ-PCR only, (K=0.3, fair agreement, p 0.08). At time -3, none resulted BCR-ABL1 positive by both methods, 4 pts were positive by ddPCR only and 1 by RQ-PCR only (K=-0.1, no agreement, p 0.62). Over one year of molecular follow-up before discontinuation, for negative values obtained measuring BCR-ABL1 by RQ-PCR the probability of relapse was equal to about 50% (45%, Figure 1), i.e. the capability of RQ-PCR of detecting relapse was basically same as random being almost constant for all the positive values of BCR-ABL1. On the contrary the probability of a relapse for negative values of BCR-ABL1 by ddPCR was about 30% and it grew up linearly at increasing level of transcript, achieving a probability of 69% when BCR-ABL1/ABL1% was equal to 0.008%. The difference between RQ-PCR and ddPCR in predicting the probability of relapse was statistically significant (p 0.02).
Conclusion
Despite the small number of cases, our results confirm that the ddPCR is more sensitive than RQ-PCR for low levels of disease. Molecular follow-up by ddPCR at several time-points before discontinuation may have a prognostic value for predicting relapse, and this is worthy to be tested prospectively and in larger cohorts of pts.

Session topic: E-poster
Keyword(s): BCR-ABL, Molecular remission, MRD, PCR
Abstract: E1090
Type: Eposter Presentation
Background
Treatment-free remission is a major goal in CML. Nowadays it is possible to safely discontinue imatinib but it is still not clear which patient (pt) will relapse. High sensitivity techniques like droplet digital (dd)PCR may help to discriminate pts who still present a significant amount of disease despite being in MR4 by standard real-time quantitative PCR (RQ-PCR).
Aims
To evaluate the capability of ddPCR to predict relapse after imatinib discontinuation in CML pts with stable MR4 by RQ-PCR.
Methods
Total RNA was extracted at different time-points from 19 pts in stable MR4 for two years before discontinuation of imatinib. ddPCR was carried out in triplicate using 200 ng of cDNA for each replicates and droplets were analyzed by QX100™ droplet reader (Bio-Rad) using the DigiDrop P210 Master Mix Kit (Bioclarma). All results were expressed according to the new recommendations [Cross, Leukemia 2015].The Wilcoxon test was used for the comparison of the total ABL medians; the Agreement Coefficient of the 2 methods was calculated with Cohen’s K test; Logistic Regression was used to assess the relationship between the probability of relapse and disease level by RQ-PCR and ddPCR.
Results
A total of 48 samples were retrospectively analyzed by RQ-PCR and ddPCR. All pts discontinued imatinib; 8/19 pts had to restart treatment for loss of MMR/MR4 after a median of 5.3 mos (2.2-8.5). A difference between ddPCR and RQ-PCR median total ABL copies (p<0.001) was found with the following medians (ranges): 46290 (8420-124600) and 93060 (25440-210000) for RQ-PCR and ddPCR respectively. A difference between ddPCR and RQ-PCR median total BCR-ABL1/ABL1% (p<0.001) was found: 0 (0-0.09); and 0.0009 (0-0.015) for RQ-PCR and ddPCR respectively. Sixteen pts had >2 evaluations, 3 had an evaluation between 14 and 18.5 mos before discontinuation. 6 pts had an evaluation at -12 mos (+/-0.81), 10 at -9 mos(+/-1.3), 14 at -6 mos(+/-0.9), 15 at -3 mos(+/-1.2), considering as time “0” the date of discontinuation. At time -12, only 1pt resulted BCR-ABL1 positive by both methods, 1 by ddPCR only and 0 by RQ-PCR only (K0.6, moderate agreement, p 0.13). At time -9, 3 pts resulted BCR-ABL1 positive by both methods, 6 by ddPCR only and none by RQ-PCR only (K=0.1, slight agreement, p 0.34). At time -6, 3 pts resulted BCR-ABL1 positive by both methods, 5 pts by ddPCR only and none by RQ-PCR only, (K=0.3, fair agreement, p 0.08). At time -3, none resulted BCR-ABL1 positive by both methods, 4 pts were positive by ddPCR only and 1 by RQ-PCR only (K=-0.1, no agreement, p 0.62). Over one year of molecular follow-up before discontinuation, for negative values obtained measuring BCR-ABL1 by RQ-PCR the probability of relapse was equal to about 50% (45%, Figure 1), i.e. the capability of RQ-PCR of detecting relapse was basically same as random being almost constant for all the positive values of BCR-ABL1. On the contrary the probability of a relapse for negative values of BCR-ABL1 by ddPCR was about 30% and it grew up linearly at increasing level of transcript, achieving a probability of 69% when BCR-ABL1/ABL1% was equal to 0.008%. The difference between RQ-PCR and ddPCR in predicting the probability of relapse was statistically significant (p 0.02).
Conclusion
Despite the small number of cases, our results confirm that the ddPCR is more sensitive than RQ-PCR for low levels of disease. Molecular follow-up by ddPCR at several time-points before discontinuation may have a prognostic value for predicting relapse, and this is worthy to be tested prospectively and in larger cohorts of pts.

Session topic: E-poster
Keyword(s): BCR-ABL, Molecular remission, MRD, PCR
Type: Eposter Presentation
Background
Treatment-free remission is a major goal in CML. Nowadays it is possible to safely discontinue imatinib but it is still not clear which patient (pt) will relapse. High sensitivity techniques like droplet digital (dd)PCR may help to discriminate pts who still present a significant amount of disease despite being in MR4 by standard real-time quantitative PCR (RQ-PCR).
Aims
To evaluate the capability of ddPCR to predict relapse after imatinib discontinuation in CML pts with stable MR4 by RQ-PCR.
Methods
Total RNA was extracted at different time-points from 19 pts in stable MR4 for two years before discontinuation of imatinib. ddPCR was carried out in triplicate using 200 ng of cDNA for each replicates and droplets were analyzed by QX100™ droplet reader (Bio-Rad) using the DigiDrop P210 Master Mix Kit (Bioclarma). All results were expressed according to the new recommendations [Cross, Leukemia 2015].The Wilcoxon test was used for the comparison of the total ABL medians; the Agreement Coefficient of the 2 methods was calculated with Cohen’s K test; Logistic Regression was used to assess the relationship between the probability of relapse and disease level by RQ-PCR and ddPCR.
Results
A total of 48 samples were retrospectively analyzed by RQ-PCR and ddPCR. All pts discontinued imatinib; 8/19 pts had to restart treatment for loss of MMR/MR4 after a median of 5.3 mos (2.2-8.5). A difference between ddPCR and RQ-PCR median total ABL copies (p<0.001) was found with the following medians (ranges): 46290 (8420-124600) and 93060 (25440-210000) for RQ-PCR and ddPCR respectively. A difference between ddPCR and RQ-PCR median total BCR-ABL1/ABL1% (p<0.001) was found: 0 (0-0.09); and 0.0009 (0-0.015) for RQ-PCR and ddPCR respectively. Sixteen pts had >2 evaluations, 3 had an evaluation between 14 and 18.5 mos before discontinuation. 6 pts had an evaluation at -12 mos (+/-0.81), 10 at -9 mos(+/-1.3), 14 at -6 mos(+/-0.9), 15 at -3 mos(+/-1.2), considering as time “0” the date of discontinuation. At time -12, only 1pt resulted BCR-ABL1 positive by both methods, 1 by ddPCR only and 0 by RQ-PCR only (K0.6, moderate agreement, p 0.13). At time -9, 3 pts resulted BCR-ABL1 positive by both methods, 6 by ddPCR only and none by RQ-PCR only (K=0.1, slight agreement, p 0.34). At time -6, 3 pts resulted BCR-ABL1 positive by both methods, 5 pts by ddPCR only and none by RQ-PCR only, (K=0.3, fair agreement, p 0.08). At time -3, none resulted BCR-ABL1 positive by both methods, 4 pts were positive by ddPCR only and 1 by RQ-PCR only (K=-0.1, no agreement, p 0.62). Over one year of molecular follow-up before discontinuation, for negative values obtained measuring BCR-ABL1 by RQ-PCR the probability of relapse was equal to about 50% (45%, Figure 1), i.e. the capability of RQ-PCR of detecting relapse was basically same as random being almost constant for all the positive values of BCR-ABL1. On the contrary the probability of a relapse for negative values of BCR-ABL1 by ddPCR was about 30% and it grew up linearly at increasing level of transcript, achieving a probability of 69% when BCR-ABL1/ABL1% was equal to 0.008%. The difference between RQ-PCR and ddPCR in predicting the probability of relapse was statistically significant (p 0.02).
Conclusion
Despite the small number of cases, our results confirm that the ddPCR is more sensitive than RQ-PCR for low levels of disease. Molecular follow-up by ddPCR at several time-points before discontinuation may have a prognostic value for predicting relapse, and this is worthy to be tested prospectively and in larger cohorts of pts.

Session topic: E-poster
Keyword(s): BCR-ABL, Molecular remission, MRD, PCR
{{ help_message }}
{{filter}}


