ARRAY COMPARATIVE GENOMIC HYBRIDIZATION HAS PROGNOSTIC VALUE IN CHRONIC LYMPHOCYTIC LEUKEMIA BEYOND STANDARD FLUORESCENCE IN SITU HYBRIDIZATION
(Abstract release date: 05/19/16)
EHA Library. Beaton B. 06/09/16; 132623; E1074

Dr. Brendan Beaton
Contributions
Contributions
Abstract
Abstract: E1074
Type: Eposter Presentation
Background
Chronic lymphocytic leukemia (CLL) has significant clinical heterogeneity. Fluorescence in situ hybridization (FISH) is an integral component of current risk stratification models, with poor prognosis associated with deletions of 11q and 17p and good prognosis with deletions at 13q. In addition to identification of clinically significant clonal aberrations by FISH, array comparative genomic hybridization (aCGH) offers a high resolution assessment of genetic imbalance.
Aims
To demonstrate prognostically significant features using aCGH beyond that shown by FISH alone.
Methods
Consent for genomic investigation on excess material was given by 91 patients (67 diagnostic, 19 post-treatment, 5 relapse) from 5 referral centres. Diagnosis was confirmed by standard morphology, immunophenotype and FISH and/or karyotype. FISH was performed using commercial probes (Abbott Molecular, UK and Cytocell, UK). aCGH was performed on DNA extracted from either peripheral blood, bone marrow aspirate, stored frozen cells or post-culture fixed chromosome preparations using a standard 8 x 60K Agilent array platform (SurePrint G3 Human CGH Microarray Kit, G4450A). Additional FISH analysis was performed to address questions raised by the microarray results when necessary. Clinical data was collected retrospectively.
Results
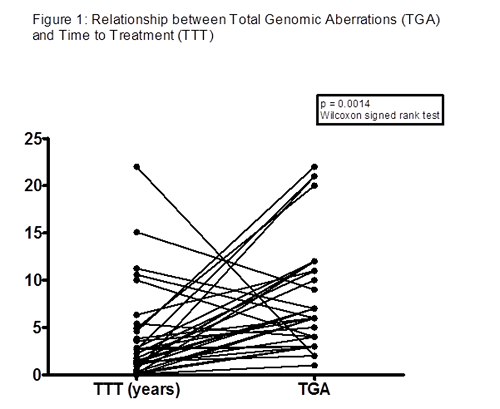
Of 91 patients (males 73.4%, females 26.6%), data regarding treatment was available for 67 (73.6%). Of these, 54 (80.5%) required chemotherapy treatment. Of those treated, median time to treatment (TTT) was 34 months (range: 0-264 months). Median time from diagnosis to end of follow-up in the untreated group was 74 months (range: 30-432 months). The known prognostic markers assessed demonstrated: diagnostic lymphocyte count >30 x109/l (73.3%); CD38 positive (48.3%); ZAP70 positive (46.5%); IGHV unmutated (43.9%); β-2 microglobulin elevated (40%). Elevated lymphocyte count and ZAP70 positivity were statistically significant between treated and untreated groups with p values of 0.023 and 0.0043, respectively. The other markers showed trends towards a difference between treated and untreated groups, but did not have a p value <0.05. There was a strong inverse relationship (see Figure 1) between TTT and total genomic aberrations (TGA) observed in the treated group (p = 0.0014).Using a cut-off of 2Mb, aCGH was able to delineate between small deletions (<2Mb) and large deletions (>2Mb) at chromosome 13q14.1. This difference cannot be delineated by standard FISH probes. Interestingly, of the untreated group, 7/13 (53.8%) had a small deletion, while 0/13 had a large deletion detected, compared with the treated group where 10/54 (18.5%) had a small deletion, while 16/54 (29.6%) had a large deletion. The difference here, where large deletions are seen in only the treated group, suggests the large deletions are associated with more aggressive disease.Another interesting observation was that by using aCGH, deletions involving Mitogen-Activated Protein Kinase Kinase 4 (MAP2K4) found at chromosome 17p, which has not yet been described in CLL, was observed in 5 patients, all in the treated group. Three of the 5 were associated with Tumor Protein P53 (TP53) found by FISH.
Conclusion
The correlation between aCGH and clinical data demonstrates the utility of aCGH beyond that of standard FISH. Increased TGA is significantly associated with shorter TTT from diagnosis. Detecting the difference between a deletion of 13q14.1 involving > 2Mb (large) compared with < 2Mb (small) by aCGH, which cannot be differentiated by standard FISH probes, may be useful in predicting patients that will need to be treated. MAP2K4 may be an important marker in CLL in predicting patients that may need treatment, but will need further investigation.
Session topic: E-poster
Keyword(s): Array based comparative genomic hybridization, Chronic lymphocytic leukemia, FISH
Type: Eposter Presentation
Background
Chronic lymphocytic leukemia (CLL) has significant clinical heterogeneity. Fluorescence in situ hybridization (FISH) is an integral component of current risk stratification models, with poor prognosis associated with deletions of 11q and 17p and good prognosis with deletions at 13q. In addition to identification of clinically significant clonal aberrations by FISH, array comparative genomic hybridization (aCGH) offers a high resolution assessment of genetic imbalance.
Aims
To demonstrate prognostically significant features using aCGH beyond that shown by FISH alone.
Methods
Consent for genomic investigation on excess material was given by 91 patients (67 diagnostic, 19 post-treatment, 5 relapse) from 5 referral centres. Diagnosis was confirmed by standard morphology, immunophenotype and FISH and/or karyotype. FISH was performed using commercial probes (Abbott Molecular, UK and Cytocell, UK). aCGH was performed on DNA extracted from either peripheral blood, bone marrow aspirate, stored frozen cells or post-culture fixed chromosome preparations using a standard 8 x 60K Agilent array platform (SurePrint G3 Human CGH Microarray Kit, G4450A). Additional FISH analysis was performed to address questions raised by the microarray results when necessary. Clinical data was collected retrospectively.
Results
Of 91 patients (males 73.4%, females 26.6%), data regarding treatment was available for 67 (73.6%). Of these, 54 (80.5%) required chemotherapy treatment. Of those treated, median time to treatment (TTT) was 34 months (range: 0-264 months). Median time from diagnosis to end of follow-up in the untreated group was 74 months (range: 30-432 months). The known prognostic markers assessed demonstrated: diagnostic lymphocyte count >30 x109/l (73.3%); CD38 positive (48.3%); ZAP70 positive (46.5%); IGHV unmutated (43.9%); β-2 microglobulin elevated (40%). Elevated lymphocyte count and ZAP70 positivity were statistically significant between treated and untreated groups with p values of 0.023 and 0.0043, respectively. The other markers showed trends towards a difference between treated and untreated groups, but did not have a p value <0.05. There was a strong inverse relationship (see Figure 1) between TTT and total genomic aberrations (TGA) observed in the treated group (p = 0.0014).Using a cut-off of 2Mb, aCGH was able to delineate between small deletions (<2Mb) and large deletions (>2Mb) at chromosome 13q14.1. This difference cannot be delineated by standard FISH probes. Interestingly, of the untreated group, 7/13 (53.8%) had a small deletion, while 0/13 had a large deletion detected, compared with the treated group where 10/54 (18.5%) had a small deletion, while 16/54 (29.6%) had a large deletion. The difference here, where large deletions are seen in only the treated group, suggests the large deletions are associated with more aggressive disease.Another interesting observation was that by using aCGH, deletions involving Mitogen-Activated Protein Kinase Kinase 4 (MAP2K4) found at chromosome 17p, which has not yet been described in CLL, was observed in 5 patients, all in the treated group. Three of the 5 were associated with Tumor Protein P53 (TP53) found by FISH.
Conclusion
The correlation between aCGH and clinical data demonstrates the utility of aCGH beyond that of standard FISH. Increased TGA is significantly associated with shorter TTT from diagnosis. Detecting the difference between a deletion of 13q14.1 involving > 2Mb (large) compared with < 2Mb (small) by aCGH, which cannot be differentiated by standard FISH probes, may be useful in predicting patients that will need to be treated. MAP2K4 may be an important marker in CLL in predicting patients that may need treatment, but will need further investigation.

Session topic: E-poster
Keyword(s): Array based comparative genomic hybridization, Chronic lymphocytic leukemia, FISH
Abstract: E1074
Type: Eposter Presentation
Background
Chronic lymphocytic leukemia (CLL) has significant clinical heterogeneity. Fluorescence in situ hybridization (FISH) is an integral component of current risk stratification models, with poor prognosis associated with deletions of 11q and 17p and good prognosis with deletions at 13q. In addition to identification of clinically significant clonal aberrations by FISH, array comparative genomic hybridization (aCGH) offers a high resolution assessment of genetic imbalance.
Aims
To demonstrate prognostically significant features using aCGH beyond that shown by FISH alone.
Methods
Consent for genomic investigation on excess material was given by 91 patients (67 diagnostic, 19 post-treatment, 5 relapse) from 5 referral centres. Diagnosis was confirmed by standard morphology, immunophenotype and FISH and/or karyotype. FISH was performed using commercial probes (Abbott Molecular, UK and Cytocell, UK). aCGH was performed on DNA extracted from either peripheral blood, bone marrow aspirate, stored frozen cells or post-culture fixed chromosome preparations using a standard 8 x 60K Agilent array platform (SurePrint G3 Human CGH Microarray Kit, G4450A). Additional FISH analysis was performed to address questions raised by the microarray results when necessary. Clinical data was collected retrospectively.
Results
Of 91 patients (males 73.4%, females 26.6%), data regarding treatment was available for 67 (73.6%). Of these, 54 (80.5%) required chemotherapy treatment. Of those treated, median time to treatment (TTT) was 34 months (range: 0-264 months). Median time from diagnosis to end of follow-up in the untreated group was 74 months (range: 30-432 months). The known prognostic markers assessed demonstrated: diagnostic lymphocyte count >30 x109/l (73.3%); CD38 positive (48.3%); ZAP70 positive (46.5%); IGHV unmutated (43.9%); β-2 microglobulin elevated (40%). Elevated lymphocyte count and ZAP70 positivity were statistically significant between treated and untreated groups with p values of 0.023 and 0.0043, respectively. The other markers showed trends towards a difference between treated and untreated groups, but did not have a p value <0.05. There was a strong inverse relationship (see Figure 1) between TTT and total genomic aberrations (TGA) observed in the treated group (p = 0.0014).Using a cut-off of 2Mb, aCGH was able to delineate between small deletions (<2Mb) and large deletions (>2Mb) at chromosome 13q14.1. This difference cannot be delineated by standard FISH probes. Interestingly, of the untreated group, 7/13 (53.8%) had a small deletion, while 0/13 had a large deletion detected, compared with the treated group where 10/54 (18.5%) had a small deletion, while 16/54 (29.6%) had a large deletion. The difference here, where large deletions are seen in only the treated group, suggests the large deletions are associated with more aggressive disease.Another interesting observation was that by using aCGH, deletions involving Mitogen-Activated Protein Kinase Kinase 4 (MAP2K4) found at chromosome 17p, which has not yet been described in CLL, was observed in 5 patients, all in the treated group. Three of the 5 were associated with Tumor Protein P53 (TP53) found by FISH.
Conclusion
The correlation between aCGH and clinical data demonstrates the utility of aCGH beyond that of standard FISH. Increased TGA is significantly associated with shorter TTT from diagnosis. Detecting the difference between a deletion of 13q14.1 involving > 2Mb (large) compared with < 2Mb (small) by aCGH, which cannot be differentiated by standard FISH probes, may be useful in predicting patients that will need to be treated. MAP2K4 may be an important marker in CLL in predicting patients that may need treatment, but will need further investigation.

Session topic: E-poster
Keyword(s): Array based comparative genomic hybridization, Chronic lymphocytic leukemia, FISH
Type: Eposter Presentation
Background
Chronic lymphocytic leukemia (CLL) has significant clinical heterogeneity. Fluorescence in situ hybridization (FISH) is an integral component of current risk stratification models, with poor prognosis associated with deletions of 11q and 17p and good prognosis with deletions at 13q. In addition to identification of clinically significant clonal aberrations by FISH, array comparative genomic hybridization (aCGH) offers a high resolution assessment of genetic imbalance.
Aims
To demonstrate prognostically significant features using aCGH beyond that shown by FISH alone.
Methods
Consent for genomic investigation on excess material was given by 91 patients (67 diagnostic, 19 post-treatment, 5 relapse) from 5 referral centres. Diagnosis was confirmed by standard morphology, immunophenotype and FISH and/or karyotype. FISH was performed using commercial probes (Abbott Molecular, UK and Cytocell, UK). aCGH was performed on DNA extracted from either peripheral blood, bone marrow aspirate, stored frozen cells or post-culture fixed chromosome preparations using a standard 8 x 60K Agilent array platform (SurePrint G3 Human CGH Microarray Kit, G4450A). Additional FISH analysis was performed to address questions raised by the microarray results when necessary. Clinical data was collected retrospectively.
Results
Of 91 patients (males 73.4%, females 26.6%), data regarding treatment was available for 67 (73.6%). Of these, 54 (80.5%) required chemotherapy treatment. Of those treated, median time to treatment (TTT) was 34 months (range: 0-264 months). Median time from diagnosis to end of follow-up in the untreated group was 74 months (range: 30-432 months). The known prognostic markers assessed demonstrated: diagnostic lymphocyte count >30 x109/l (73.3%); CD38 positive (48.3%); ZAP70 positive (46.5%); IGHV unmutated (43.9%); β-2 microglobulin elevated (40%). Elevated lymphocyte count and ZAP70 positivity were statistically significant between treated and untreated groups with p values of 0.023 and 0.0043, respectively. The other markers showed trends towards a difference between treated and untreated groups, but did not have a p value <0.05. There was a strong inverse relationship (see Figure 1) between TTT and total genomic aberrations (TGA) observed in the treated group (p = 0.0014).Using a cut-off of 2Mb, aCGH was able to delineate between small deletions (<2Mb) and large deletions (>2Mb) at chromosome 13q14.1. This difference cannot be delineated by standard FISH probes. Interestingly, of the untreated group, 7/13 (53.8%) had a small deletion, while 0/13 had a large deletion detected, compared with the treated group where 10/54 (18.5%) had a small deletion, while 16/54 (29.6%) had a large deletion. The difference here, where large deletions are seen in only the treated group, suggests the large deletions are associated with more aggressive disease.Another interesting observation was that by using aCGH, deletions involving Mitogen-Activated Protein Kinase Kinase 4 (MAP2K4) found at chromosome 17p, which has not yet been described in CLL, was observed in 5 patients, all in the treated group. Three of the 5 were associated with Tumor Protein P53 (TP53) found by FISH.
Conclusion
The correlation between aCGH and clinical data demonstrates the utility of aCGH beyond that of standard FISH. Increased TGA is significantly associated with shorter TTT from diagnosis. Detecting the difference between a deletion of 13q14.1 involving > 2Mb (large) compared with < 2Mb (small) by aCGH, which cannot be differentiated by standard FISH probes, may be useful in predicting patients that will need to be treated. MAP2K4 may be an important marker in CLL in predicting patients that may need treatment, but will need further investigation.

Session topic: E-poster
Keyword(s): Array based comparative genomic hybridization, Chronic lymphocytic leukemia, FISH
{{ help_message }}
{{filter}}


