SF3B1 MUTATIONS, IGHV MUTATION STATUS AND ABNORMALITIES DETECTED BY GENOMIC ARRAYS DETERMINE PROGNOSIS IN CLL WITH NORMAL KARYOTYPE
(Abstract release date: 05/19/16)
EHA Library. Vetro C. 06/09/16; 132618; E1069

Mr. Calogero Vetro
Contributions
Contributions
Abstract
Abstract: E1069
Type: Eposter Presentation
Background
Normal karyotype (NK) CLL accounts for 15-20% of all CLL cases and confers per se an intermediate risk. Our group reported a higher SF3B1 mutation (mut) rate compared to other cytogenetic subgroups and only one third of patients showed unmutated IGHV status (IGHVu). Furthermore, we previously demonstrated that array-based comparative genomic hybridization (aCGH) technology could detect cryptic abnormalities in 19% of NK CLL.
Aims
To further characterize CLL with normal karyotype and identify prognostic parameters.
Methods
We selected 164 patients with NK and CLL cells percentage >15% (16% of total CLL cohort). Chromosome banding analysis yielded normal karyotypes. FISH analyses, performed with probes for 17p13 (TP53), 13q14 (D13S25, D13S319, DLEU), 11q22 (ATM), centromere region of chromosome 12, t(11;14)(q13;q32)(IGH-CCND1), 6q21 and IGH rearrangements, did not reveal any abnormalities. aCGH was performed with SurePrint G3 ISCA CGH+SNP Microarray (Agilent, Waldbronn, Germany). IGHV status, mutational analysis by DNA sequencing for SF3B1, NOTCH1 and TP53 and information on time to treatment (TTT) were available for all patients. White blood cell (WBC) count was available for 153/164 patients.
Results
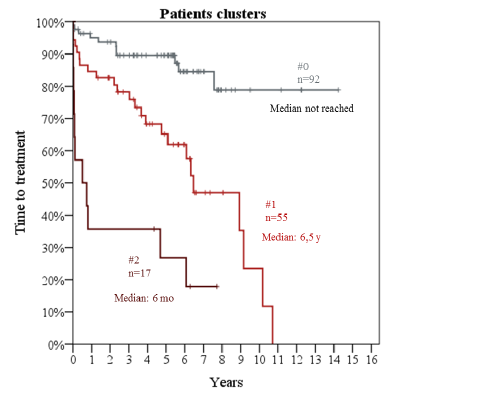
Median follow-up was 5.6 years and 10-year OS was 75%. Median age was 63 years (range, 34 to 83) and 93/164 (54%) were male. Median WBC count was 36,500/mL (range, 3,300 to 463,000). Cases with CLL cells ≤ 5000/μL had nodal presentation. By flow cytometry, median % CLL cells was 52% (range, 16% to 94%). Cut-off for CD38 and ZAP70 positivity was 30% and 20%, respectively. 34 patients (20%) showed CD38 positivity and 61 (36%) ZAP70 positivity. By aCGH, we detected 68 copy number abnormalities (CNA) in 37 patients (22%). Majority (24/37; 65%) had 1 CNA, 9 patients (24%) showed 2 CNA and 4 (11%) more than 2 CNA. IGHVu was present in 52/164 (32%) cases. SF3B1 was mutated in 26/164 (16%), NOTCH1 in 12 (7%) and TP53 in 7(4%). Univariate Cox analysis for TTT showed adverse impact for: WBC (HR:1.05 per 10,000/μL increase; 95%CI 1.02-1.09; p=0.001); % CLL cells (HR:1.3 per 10% increase; 95%CI 1.16-1.5; p<0.001); CD38 positivity (HR:2.7; 95%CI 1.5-5; p=0.001); ≥2 CNA (HR:3.6; 95%CI 1.6-8.1; p=0.002); SF3B1mut (HR:2.3; 95%CI 1.2-4.5 p=0.013); IGHVu (HR:4.8; 95%CI 2.6-8.7; p<0.001). Detection of any CNA, ZAP70 positivity, TP53mut and NOTCH1mut did not relate with TTT. In multivariate analysis, % CLL cells (HR:1.2 per 10% increase; 95%CI 1.04-1.4; p=0.015), ≥2 CNA (HR:2.9; 95%CI 1.2-7.012; p=0.015), SF3B1mut (HR:2.3; 95%CI 1.16-4.5; p=0.016) and IGHVu (HR:4.3; 95%CI 2.1-8.4; p<0.001) retained their prognostic value. Therefore, we clustered patients into 3 groups (figure 1; table 1). Patients without risk factors (#0) showed the best outcome (median TTT not reached); patients with 1 risk factor (#1) had a median TTT of 6.5 years; patients with at least two risk factors (#2) showed a median TTT of 6 months (p<0.001).Table 1. Cluster groups according to prognostic factors
Conclusion
Through the evaluation of IGHV mutational status, SF3B1 and aCGH, we identified CLL patients with normal karyotype at the highest risk for need of treatment.
Session topic: E-poster
Keyword(s): Array based comparative genomic hybridization, Chronic lymphocytic leukemia, Mutation, Prognosis
Type: Eposter Presentation
Background
Normal karyotype (NK) CLL accounts for 15-20% of all CLL cases and confers per se an intermediate risk. Our group reported a higher SF3B1 mutation (mut) rate compared to other cytogenetic subgroups and only one third of patients showed unmutated IGHV status (IGHVu). Furthermore, we previously demonstrated that array-based comparative genomic hybridization (aCGH) technology could detect cryptic abnormalities in 19% of NK CLL.
Aims
To further characterize CLL with normal karyotype and identify prognostic parameters.
Methods
We selected 164 patients with NK and CLL cells percentage >15% (16% of total CLL cohort). Chromosome banding analysis yielded normal karyotypes. FISH analyses, performed with probes for 17p13 (TP53), 13q14 (D13S25, D13S319, DLEU), 11q22 (ATM), centromere region of chromosome 12, t(11;14)(q13;q32)(IGH-CCND1), 6q21 and IGH rearrangements, did not reveal any abnormalities. aCGH was performed with SurePrint G3 ISCA CGH+SNP Microarray (Agilent, Waldbronn, Germany). IGHV status, mutational analysis by DNA sequencing for SF3B1, NOTCH1 and TP53 and information on time to treatment (TTT) were available for all patients. White blood cell (WBC) count was available for 153/164 patients.
Results
Median follow-up was 5.6 years and 10-year OS was 75%. Median age was 63 years (range, 34 to 83) and 93/164 (54%) were male. Median WBC count was 36,500/mL (range, 3,300 to 463,000). Cases with CLL cells ≤ 5000/μL had nodal presentation. By flow cytometry, median % CLL cells was 52% (range, 16% to 94%). Cut-off for CD38 and ZAP70 positivity was 30% and 20%, respectively. 34 patients (20%) showed CD38 positivity and 61 (36%) ZAP70 positivity. By aCGH, we detected 68 copy number abnormalities (CNA) in 37 patients (22%). Majority (24/37; 65%) had 1 CNA, 9 patients (24%) showed 2 CNA and 4 (11%) more than 2 CNA. IGHVu was present in 52/164 (32%) cases. SF3B1 was mutated in 26/164 (16%), NOTCH1 in 12 (7%) and TP53 in 7(4%). Univariate Cox analysis for TTT showed adverse impact for: WBC (HR:1.05 per 10,000/μL increase; 95%CI 1.02-1.09; p=0.001); % CLL cells (HR:1.3 per 10% increase; 95%CI 1.16-1.5; p<0.001); CD38 positivity (HR:2.7; 95%CI 1.5-5; p=0.001); ≥2 CNA (HR:3.6; 95%CI 1.6-8.1; p=0.002); SF3B1mut (HR:2.3; 95%CI 1.2-4.5 p=0.013); IGHVu (HR:4.8; 95%CI 2.6-8.7; p<0.001). Detection of any CNA, ZAP70 positivity, TP53mut and NOTCH1mut did not relate with TTT. In multivariate analysis, % CLL cells (HR:1.2 per 10% increase; 95%CI 1.04-1.4; p=0.015), ≥2 CNA (HR:2.9; 95%CI 1.2-7.012; p=0.015), SF3B1mut (HR:2.3; 95%CI 1.16-4.5; p=0.016) and IGHVu (HR:4.3; 95%CI 2.1-8.4; p<0.001) retained their prognostic value. Therefore, we clustered patients into 3 groups (figure 1; table 1). Patients without risk factors (#0) showed the best outcome (median TTT not reached); patients with 1 risk factor (#1) had a median TTT of 6.5 years; patients with at least two risk factors (#2) showed a median TTT of 6 months (p<0.001).
| Cluster | number of cases | Prognostic factors | median TTT | |
| #0 | 92 | no | Not reached | |
| #1 | 55 | 15 | SF3B1mut | 6.6 yrs |
| 35 | IGHVu | |||
| 5 | CNA≥2 | |||
| #2 | 17 | 9 | SF3B1mut AND IGHVu | 6 mo |
| 6 | IGHVu AND CNA≥2 | |||
| 2 | IGHVu AND SF3B1mut AND CNA≥2 | |||
Conclusion
Through the evaluation of IGHV mutational status, SF3B1 and aCGH, we identified CLL patients with normal karyotype at the highest risk for need of treatment.

Session topic: E-poster
Keyword(s): Array based comparative genomic hybridization, Chronic lymphocytic leukemia, Mutation, Prognosis
Abstract: E1069
Type: Eposter Presentation
Background
Normal karyotype (NK) CLL accounts for 15-20% of all CLL cases and confers per se an intermediate risk. Our group reported a higher SF3B1 mutation (mut) rate compared to other cytogenetic subgroups and only one third of patients showed unmutated IGHV status (IGHVu). Furthermore, we previously demonstrated that array-based comparative genomic hybridization (aCGH) technology could detect cryptic abnormalities in 19% of NK CLL.
Aims
To further characterize CLL with normal karyotype and identify prognostic parameters.
Methods
We selected 164 patients with NK and CLL cells percentage >15% (16% of total CLL cohort). Chromosome banding analysis yielded normal karyotypes. FISH analyses, performed with probes for 17p13 (TP53), 13q14 (D13S25, D13S319, DLEU), 11q22 (ATM), centromere region of chromosome 12, t(11;14)(q13;q32)(IGH-CCND1), 6q21 and IGH rearrangements, did not reveal any abnormalities. aCGH was performed with SurePrint G3 ISCA CGH+SNP Microarray (Agilent, Waldbronn, Germany). IGHV status, mutational analysis by DNA sequencing for SF3B1, NOTCH1 and TP53 and information on time to treatment (TTT) were available for all patients. White blood cell (WBC) count was available for 153/164 patients.
Results
Median follow-up was 5.6 years and 10-year OS was 75%. Median age was 63 years (range, 34 to 83) and 93/164 (54%) were male. Median WBC count was 36,500/mL (range, 3,300 to 463,000). Cases with CLL cells ≤ 5000/μL had nodal presentation. By flow cytometry, median % CLL cells was 52% (range, 16% to 94%). Cut-off for CD38 and ZAP70 positivity was 30% and 20%, respectively. 34 patients (20%) showed CD38 positivity and 61 (36%) ZAP70 positivity. By aCGH, we detected 68 copy number abnormalities (CNA) in 37 patients (22%). Majority (24/37; 65%) had 1 CNA, 9 patients (24%) showed 2 CNA and 4 (11%) more than 2 CNA. IGHVu was present in 52/164 (32%) cases. SF3B1 was mutated in 26/164 (16%), NOTCH1 in 12 (7%) and TP53 in 7(4%). Univariate Cox analysis for TTT showed adverse impact for: WBC (HR:1.05 per 10,000/μL increase; 95%CI 1.02-1.09; p=0.001); % CLL cells (HR:1.3 per 10% increase; 95%CI 1.16-1.5; p<0.001); CD38 positivity (HR:2.7; 95%CI 1.5-5; p=0.001); ≥2 CNA (HR:3.6; 95%CI 1.6-8.1; p=0.002); SF3B1mut (HR:2.3; 95%CI 1.2-4.5 p=0.013); IGHVu (HR:4.8; 95%CI 2.6-8.7; p<0.001). Detection of any CNA, ZAP70 positivity, TP53mut and NOTCH1mut did not relate with TTT. In multivariate analysis, % CLL cells (HR:1.2 per 10% increase; 95%CI 1.04-1.4; p=0.015), ≥2 CNA (HR:2.9; 95%CI 1.2-7.012; p=0.015), SF3B1mut (HR:2.3; 95%CI 1.16-4.5; p=0.016) and IGHVu (HR:4.3; 95%CI 2.1-8.4; p<0.001) retained their prognostic value. Therefore, we clustered patients into 3 groups (figure 1; table 1). Patients without risk factors (#0) showed the best outcome (median TTT not reached); patients with 1 risk factor (#1) had a median TTT of 6.5 years; patients with at least two risk factors (#2) showed a median TTT of 6 months (p<0.001).Table 1. Cluster groups according to prognostic factors
Conclusion
Through the evaluation of IGHV mutational status, SF3B1 and aCGH, we identified CLL patients with normal karyotype at the highest risk for need of treatment.

Session topic: E-poster
Keyword(s): Array based comparative genomic hybridization, Chronic lymphocytic leukemia, Mutation, Prognosis
Type: Eposter Presentation
Background
Normal karyotype (NK) CLL accounts for 15-20% of all CLL cases and confers per se an intermediate risk. Our group reported a higher SF3B1 mutation (mut) rate compared to other cytogenetic subgroups and only one third of patients showed unmutated IGHV status (IGHVu). Furthermore, we previously demonstrated that array-based comparative genomic hybridization (aCGH) technology could detect cryptic abnormalities in 19% of NK CLL.
Aims
To further characterize CLL with normal karyotype and identify prognostic parameters.
Methods
We selected 164 patients with NK and CLL cells percentage >15% (16% of total CLL cohort). Chromosome banding analysis yielded normal karyotypes. FISH analyses, performed with probes for 17p13 (TP53), 13q14 (D13S25, D13S319, DLEU), 11q22 (ATM), centromere region of chromosome 12, t(11;14)(q13;q32)(IGH-CCND1), 6q21 and IGH rearrangements, did not reveal any abnormalities. aCGH was performed with SurePrint G3 ISCA CGH+SNP Microarray (Agilent, Waldbronn, Germany). IGHV status, mutational analysis by DNA sequencing for SF3B1, NOTCH1 and TP53 and information on time to treatment (TTT) were available for all patients. White blood cell (WBC) count was available for 153/164 patients.
Results
Median follow-up was 5.6 years and 10-year OS was 75%. Median age was 63 years (range, 34 to 83) and 93/164 (54%) were male. Median WBC count was 36,500/mL (range, 3,300 to 463,000). Cases with CLL cells ≤ 5000/μL had nodal presentation. By flow cytometry, median % CLL cells was 52% (range, 16% to 94%). Cut-off for CD38 and ZAP70 positivity was 30% and 20%, respectively. 34 patients (20%) showed CD38 positivity and 61 (36%) ZAP70 positivity. By aCGH, we detected 68 copy number abnormalities (CNA) in 37 patients (22%). Majority (24/37; 65%) had 1 CNA, 9 patients (24%) showed 2 CNA and 4 (11%) more than 2 CNA. IGHVu was present in 52/164 (32%) cases. SF3B1 was mutated in 26/164 (16%), NOTCH1 in 12 (7%) and TP53 in 7(4%). Univariate Cox analysis for TTT showed adverse impact for: WBC (HR:1.05 per 10,000/μL increase; 95%CI 1.02-1.09; p=0.001); % CLL cells (HR:1.3 per 10% increase; 95%CI 1.16-1.5; p<0.001); CD38 positivity (HR:2.7; 95%CI 1.5-5; p=0.001); ≥2 CNA (HR:3.6; 95%CI 1.6-8.1; p=0.002); SF3B1mut (HR:2.3; 95%CI 1.2-4.5 p=0.013); IGHVu (HR:4.8; 95%CI 2.6-8.7; p<0.001). Detection of any CNA, ZAP70 positivity, TP53mut and NOTCH1mut did not relate with TTT. In multivariate analysis, % CLL cells (HR:1.2 per 10% increase; 95%CI 1.04-1.4; p=0.015), ≥2 CNA (HR:2.9; 95%CI 1.2-7.012; p=0.015), SF3B1mut (HR:2.3; 95%CI 1.16-4.5; p=0.016) and IGHVu (HR:4.3; 95%CI 2.1-8.4; p<0.001) retained their prognostic value. Therefore, we clustered patients into 3 groups (figure 1; table 1). Patients without risk factors (#0) showed the best outcome (median TTT not reached); patients with 1 risk factor (#1) had a median TTT of 6.5 years; patients with at least two risk factors (#2) showed a median TTT of 6 months (p<0.001).
| Cluster | number of cases | Prognostic factors | median TTT | |
| #0 | 92 | no | Not reached | |
| #1 | 55 | 15 | SF3B1mut | 6.6 yrs |
| 35 | IGHVu | |||
| 5 | CNA≥2 | |||
| #2 | 17 | 9 | SF3B1mut AND IGHVu | 6 mo |
| 6 | IGHVu AND CNA≥2 | |||
| 2 | IGHVu AND SF3B1mut AND CNA≥2 | |||
Conclusion
Through the evaluation of IGHV mutational status, SF3B1 and aCGH, we identified CLL patients with normal karyotype at the highest risk for need of treatment.

Session topic: E-poster
Keyword(s): Array based comparative genomic hybridization, Chronic lymphocytic leukemia, Mutation, Prognosis
{{ help_message }}
{{filter}}


