INCIDENTAL FINDINGS BY TARGETED EXOME SEQUENCING USED TO DIAGNOSE RARE INHERITED BLEEDING DISORDERS – PATIENT`S CHOICES ON INFORMATION AND OUTCOME.
(Abstract release date: 05/19/16)
EHA Library. Leinoe E. 06/09/16; 132537; E988

Dr. Eva Leinoe
Contributions
Contributions
Abstract
Abstract: E988
Type: Eposter Presentation
Background
Whole exome sequencing (WES) has the ability to identify numerous mutations within a genome, many of which are not related to the phenotype in question, but they may have clinical ramifications. Incidental findings (IF) are pathogenic alterations in genes, that are not apparently relevant to the diagnostic situation, for which, the sequencing was intended. Scientific and ethics boards recommend, that the informed consent process ensures, that patients understand the possibility that IF will be detected and indicate which (if any) information on IF, they wish to receive. We have implemented WES to diagnose patients with inherited bleeding disorders. Our exome sequencing panel for bleeding disorders only examines specific genes, but with the additional use of search terms (thrombocytopenia, thrombocytopathia and bleeding) incidental findings may occur. Mutations associated with inherited thrombocytopenia may also be associated with haematological cancers. No previous study has described the outcome of IF in this diagnostic setting.
Aims
To examine patient`s choice on information regarding IF and to report the rate of IF.
Methods
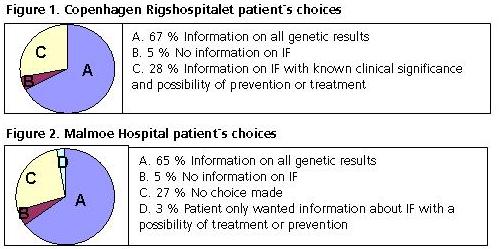
Patients were examined by ISTH-BAT score and standard coagulation testing. Prior to WES analysis, patients were given oral and written information concerning the risk and possible seriousness of IF. The pretest information on IF included risk of detecting mutations associated with cancer or brain disease. Patients from Copenhagen were asked to make a pretest written choice of whether they wanted information on: A. All genetic results of clinical significance, including IF. B. No information on IF. C. Provision of IF with known clinical significance and the possibility of treatment or prevention of disease. Patients from Malmoe were asked to make a pretest written choice of whether they wanted information on: A. All genetic results of clinical significance, including IF. B. No information on IF. The Illumina HiSeq 2500 platform was used for targeted exome sequencing. Sequencing was performed on the HiSeq 2500 as paired end (PE) sequencing, 2x101 bases, resulting in approximately 100 M PE reads. Variants were called with a minimum of 10x coverage and exported as .vcf format with approximately 220,000 variants prior to upload in Ingenuity Variant Analysis (Qiagen). Common SNPs were subtracted.
Results
A total of 91 genes associated with bleeding were examined in 107 patients from Copenhagen and 37 patients from Malmoe. The following IF were made: 1. A heterozygous variant in Telomerase reverse transcriptase (TERT) (His412Tyr) in a 34-year female with macro-thrombocytopenia. The variant had previously been reported to be associated with reduced telomerase activity, dyskeratosis congenital and aplastic anemia. A telomer analysis performed by an outside lab, showed that leukocytes did not have short telomeres or low telomerase enzymatic activity. It was concluded, that the variant was a nonpathogenic polymorphism and the patient informed correspondently. 2. A heterozygous missense mutation in SERPINC1 (Pro73Leu) in a 40-year female without thrombosis. It was previously described as a founder mutation in the Finnish population, associated with type II antithrombin (AT) deficiency and increased risk of thrombosis. Her AT III level was normal; no additional function testing was done. It was concluded that due to her haemorrhagic diathesis, she was not at high risk of thrombosis and informed correspondently. 3. A heterozygous GATA2 mutation (Pro41Ala) previously associated with myelodysplasia and skin cancer in a 36-year female. Several relatives had a history of skin cancer and her aunt had leukemia. She was informed of the increased risk of skin and haematological cancer, offered regular follow-up and advised on skin protection.
Conclusion
IF were discovered in 3/148 (2%). In two separate cohorts of patients from Copenhagen and Malmoe, the choices on information were comparable. Ninety-nine patients (66%) wanted information on all genetic results and 7 patients (5%) declined all information regarding IF. The remaining 42 patients (29%) did either not make a choice or only wanted information, when a disease could be treated or prevented. Targeted exome sequencing of rare inherited bleeding disorders may disclose IF with major impact and/or require additional consultation/validation by an external laboratory or expert. It is of outmost importance that patients understand the consequences of IF, before giving informed consent to WES:
Session topic: E-poster
Keyword(s): Bleeding disorder, Diagnosis, Genomics, Thrombocytopenia
Type: Eposter Presentation
Background
Whole exome sequencing (WES) has the ability to identify numerous mutations within a genome, many of which are not related to the phenotype in question, but they may have clinical ramifications. Incidental findings (IF) are pathogenic alterations in genes, that are not apparently relevant to the diagnostic situation, for which, the sequencing was intended. Scientific and ethics boards recommend, that the informed consent process ensures, that patients understand the possibility that IF will be detected and indicate which (if any) information on IF, they wish to receive. We have implemented WES to diagnose patients with inherited bleeding disorders. Our exome sequencing panel for bleeding disorders only examines specific genes, but with the additional use of search terms (thrombocytopenia, thrombocytopathia and bleeding) incidental findings may occur. Mutations associated with inherited thrombocytopenia may also be associated with haematological cancers. No previous study has described the outcome of IF in this diagnostic setting.
Aims
To examine patient`s choice on information regarding IF and to report the rate of IF.
Methods
Patients were examined by ISTH-BAT score and standard coagulation testing. Prior to WES analysis, patients were given oral and written information concerning the risk and possible seriousness of IF. The pretest information on IF included risk of detecting mutations associated with cancer or brain disease. Patients from Copenhagen were asked to make a pretest written choice of whether they wanted information on: A. All genetic results of clinical significance, including IF. B. No information on IF. C. Provision of IF with known clinical significance and the possibility of treatment or prevention of disease. Patients from Malmoe were asked to make a pretest written choice of whether they wanted information on: A. All genetic results of clinical significance, including IF. B. No information on IF. The Illumina HiSeq 2500 platform was used for targeted exome sequencing. Sequencing was performed on the HiSeq 2500 as paired end (PE) sequencing, 2x101 bases, resulting in approximately 100 M PE reads. Variants were called with a minimum of 10x coverage and exported as .vcf format with approximately 220,000 variants prior to upload in Ingenuity Variant Analysis (Qiagen). Common SNPs were subtracted.
Results
A total of 91 genes associated with bleeding were examined in 107 patients from Copenhagen and 37 patients from Malmoe. The following IF were made: 1. A heterozygous variant in Telomerase reverse transcriptase (TERT) (His412Tyr) in a 34-year female with macro-thrombocytopenia. The variant had previously been reported to be associated with reduced telomerase activity, dyskeratosis congenital and aplastic anemia. A telomer analysis performed by an outside lab, showed that leukocytes did not have short telomeres or low telomerase enzymatic activity. It was concluded, that the variant was a nonpathogenic polymorphism and the patient informed correspondently. 2. A heterozygous missense mutation in SERPINC1 (Pro73Leu) in a 40-year female without thrombosis. It was previously described as a founder mutation in the Finnish population, associated with type II antithrombin (AT) deficiency and increased risk of thrombosis. Her AT III level was normal; no additional function testing was done. It was concluded that due to her haemorrhagic diathesis, she was not at high risk of thrombosis and informed correspondently. 3. A heterozygous GATA2 mutation (Pro41Ala) previously associated with myelodysplasia and skin cancer in a 36-year female. Several relatives had a history of skin cancer and her aunt had leukemia. She was informed of the increased risk of skin and haematological cancer, offered regular follow-up and advised on skin protection.
Conclusion
IF were discovered in 3/148 (2%). In two separate cohorts of patients from Copenhagen and Malmoe, the choices on information were comparable. Ninety-nine patients (66%) wanted information on all genetic results and 7 patients (5%) declined all information regarding IF. The remaining 42 patients (29%) did either not make a choice or only wanted information, when a disease could be treated or prevented. Targeted exome sequencing of rare inherited bleeding disorders may disclose IF with major impact and/or require additional consultation/validation by an external laboratory or expert. It is of outmost importance that patients understand the consequences of IF, before giving informed consent to WES:

Session topic: E-poster
Keyword(s): Bleeding disorder, Diagnosis, Genomics, Thrombocytopenia
Abstract: E988
Type: Eposter Presentation
Background
Whole exome sequencing (WES) has the ability to identify numerous mutations within a genome, many of which are not related to the phenotype in question, but they may have clinical ramifications. Incidental findings (IF) are pathogenic alterations in genes, that are not apparently relevant to the diagnostic situation, for which, the sequencing was intended. Scientific and ethics boards recommend, that the informed consent process ensures, that patients understand the possibility that IF will be detected and indicate which (if any) information on IF, they wish to receive. We have implemented WES to diagnose patients with inherited bleeding disorders. Our exome sequencing panel for bleeding disorders only examines specific genes, but with the additional use of search terms (thrombocytopenia, thrombocytopathia and bleeding) incidental findings may occur. Mutations associated with inherited thrombocytopenia may also be associated with haematological cancers. No previous study has described the outcome of IF in this diagnostic setting.
Aims
To examine patient`s choice on information regarding IF and to report the rate of IF.
Methods
Patients were examined by ISTH-BAT score and standard coagulation testing. Prior to WES analysis, patients were given oral and written information concerning the risk and possible seriousness of IF. The pretest information on IF included risk of detecting mutations associated with cancer or brain disease. Patients from Copenhagen were asked to make a pretest written choice of whether they wanted information on: A. All genetic results of clinical significance, including IF. B. No information on IF. C. Provision of IF with known clinical significance and the possibility of treatment or prevention of disease. Patients from Malmoe were asked to make a pretest written choice of whether they wanted information on: A. All genetic results of clinical significance, including IF. B. No information on IF. The Illumina HiSeq 2500 platform was used for targeted exome sequencing. Sequencing was performed on the HiSeq 2500 as paired end (PE) sequencing, 2x101 bases, resulting in approximately 100 M PE reads. Variants were called with a minimum of 10x coverage and exported as .vcf format with approximately 220,000 variants prior to upload in Ingenuity Variant Analysis (Qiagen). Common SNPs were subtracted.
Results
A total of 91 genes associated with bleeding were examined in 107 patients from Copenhagen and 37 patients from Malmoe. The following IF were made: 1. A heterozygous variant in Telomerase reverse transcriptase (TERT) (His412Tyr) in a 34-year female with macro-thrombocytopenia. The variant had previously been reported to be associated with reduced telomerase activity, dyskeratosis congenital and aplastic anemia. A telomer analysis performed by an outside lab, showed that leukocytes did not have short telomeres or low telomerase enzymatic activity. It was concluded, that the variant was a nonpathogenic polymorphism and the patient informed correspondently. 2. A heterozygous missense mutation in SERPINC1 (Pro73Leu) in a 40-year female without thrombosis. It was previously described as a founder mutation in the Finnish population, associated with type II antithrombin (AT) deficiency and increased risk of thrombosis. Her AT III level was normal; no additional function testing was done. It was concluded that due to her haemorrhagic diathesis, she was not at high risk of thrombosis and informed correspondently. 3. A heterozygous GATA2 mutation (Pro41Ala) previously associated with myelodysplasia and skin cancer in a 36-year female. Several relatives had a history of skin cancer and her aunt had leukemia. She was informed of the increased risk of skin and haematological cancer, offered regular follow-up and advised on skin protection.
Conclusion
IF were discovered in 3/148 (2%). In two separate cohorts of patients from Copenhagen and Malmoe, the choices on information were comparable. Ninety-nine patients (66%) wanted information on all genetic results and 7 patients (5%) declined all information regarding IF. The remaining 42 patients (29%) did either not make a choice or only wanted information, when a disease could be treated or prevented. Targeted exome sequencing of rare inherited bleeding disorders may disclose IF with major impact and/or require additional consultation/validation by an external laboratory or expert. It is of outmost importance that patients understand the consequences of IF, before giving informed consent to WES:

Session topic: E-poster
Keyword(s): Bleeding disorder, Diagnosis, Genomics, Thrombocytopenia
Type: Eposter Presentation
Background
Whole exome sequencing (WES) has the ability to identify numerous mutations within a genome, many of which are not related to the phenotype in question, but they may have clinical ramifications. Incidental findings (IF) are pathogenic alterations in genes, that are not apparently relevant to the diagnostic situation, for which, the sequencing was intended. Scientific and ethics boards recommend, that the informed consent process ensures, that patients understand the possibility that IF will be detected and indicate which (if any) information on IF, they wish to receive. We have implemented WES to diagnose patients with inherited bleeding disorders. Our exome sequencing panel for bleeding disorders only examines specific genes, but with the additional use of search terms (thrombocytopenia, thrombocytopathia and bleeding) incidental findings may occur. Mutations associated with inherited thrombocytopenia may also be associated with haematological cancers. No previous study has described the outcome of IF in this diagnostic setting.
Aims
To examine patient`s choice on information regarding IF and to report the rate of IF.
Methods
Patients were examined by ISTH-BAT score and standard coagulation testing. Prior to WES analysis, patients were given oral and written information concerning the risk and possible seriousness of IF. The pretest information on IF included risk of detecting mutations associated with cancer or brain disease. Patients from Copenhagen were asked to make a pretest written choice of whether they wanted information on: A. All genetic results of clinical significance, including IF. B. No information on IF. C. Provision of IF with known clinical significance and the possibility of treatment or prevention of disease. Patients from Malmoe were asked to make a pretest written choice of whether they wanted information on: A. All genetic results of clinical significance, including IF. B. No information on IF. The Illumina HiSeq 2500 platform was used for targeted exome sequencing. Sequencing was performed on the HiSeq 2500 as paired end (PE) sequencing, 2x101 bases, resulting in approximately 100 M PE reads. Variants were called with a minimum of 10x coverage and exported as .vcf format with approximately 220,000 variants prior to upload in Ingenuity Variant Analysis (Qiagen). Common SNPs were subtracted.
Results
A total of 91 genes associated with bleeding were examined in 107 patients from Copenhagen and 37 patients from Malmoe. The following IF were made: 1. A heterozygous variant in Telomerase reverse transcriptase (TERT) (His412Tyr) in a 34-year female with macro-thrombocytopenia. The variant had previously been reported to be associated with reduced telomerase activity, dyskeratosis congenital and aplastic anemia. A telomer analysis performed by an outside lab, showed that leukocytes did not have short telomeres or low telomerase enzymatic activity. It was concluded, that the variant was a nonpathogenic polymorphism and the patient informed correspondently. 2. A heterozygous missense mutation in SERPINC1 (Pro73Leu) in a 40-year female without thrombosis. It was previously described as a founder mutation in the Finnish population, associated with type II antithrombin (AT) deficiency and increased risk of thrombosis. Her AT III level was normal; no additional function testing was done. It was concluded that due to her haemorrhagic diathesis, she was not at high risk of thrombosis and informed correspondently. 3. A heterozygous GATA2 mutation (Pro41Ala) previously associated with myelodysplasia and skin cancer in a 36-year female. Several relatives had a history of skin cancer and her aunt had leukemia. She was informed of the increased risk of skin and haematological cancer, offered regular follow-up and advised on skin protection.
Conclusion
IF were discovered in 3/148 (2%). In two separate cohorts of patients from Copenhagen and Malmoe, the choices on information were comparable. Ninety-nine patients (66%) wanted information on all genetic results and 7 patients (5%) declined all information regarding IF. The remaining 42 patients (29%) did either not make a choice or only wanted information, when a disease could be treated or prevented. Targeted exome sequencing of rare inherited bleeding disorders may disclose IF with major impact and/or require additional consultation/validation by an external laboratory or expert. It is of outmost importance that patients understand the consequences of IF, before giving informed consent to WES:

Session topic: E-poster
Keyword(s): Bleeding disorder, Diagnosis, Genomics, Thrombocytopenia
{{ help_message }}
{{filter}}


