A SIMPLE 3-GENE MOLECULAR SCORE RETAINS HIGH PROGNOSTIC VALUE IN NON HIGH RISK ACUTE PROMYELOCYTIC LEUKEMIA TREATED WITH AIDA 2000 PROTOCOL: A RETROSPECTIVE STUDY FROM A REGIONAL REGISTER
(Abstract release date: 05/19/16)
EHA Library. Guardo D. 06/09/16; 132470; E921

Dr. Daniela Guardo
Contributions
Contributions
Abstract
Abstract: E921
Type: Eposter Presentation
Background
Non high risk acute promyelocytic leukemia (APL) patients, as defined by WBC count at diagnosis, have nowadays a very good prognosis when treated with ATRA plus chemotherapy based protocols, with high rate of complete molecular remission after consolidation therapy. However, a small portion of patients (roughly 10%) will eventually relapse, despite the achievement of molecular CR.In the past years, some groups showed that relapse risk could be predicted by testing for some gene alterations, such as FLT3-ITD, or by break point cluster region (BCR) analysis. However, those results were not confirmed in prospective trials.
Aims
We recently showed that a simple gene-expression panel including WT1 and BAALC expression levels analysis has high prognostic value in MDS patients. The aim of the present study was to evaluate if the application of a similar panel to APL patients at diagnosis could be predictive of relapse risk and survival.
Methods
We retrospectively applied a simple 3 gene based molecular panel including WT1 and BAALC expression levels at diagnosis, FLT3-ITD mutational status (molecular profile), alongside with BCR analysis, to a cohort of de novo 66 non high-risk acute promyelocytic leukemia patients, treated in our institution between January 1st 2004 to June 30th 2015. All patients have been treated according to the Italian age-adapted AIDA protocol. Median age was 47 years (range 16-88), 17 patients were older than 60 years. Median follow up was 58 months.BCR analysis was available in all patients, 43 patients showed BCR1/2, whereas 23 patients had BCR3 (35%).Molecular profile was available in 44/66 patients (66%). Cut-off values for WT1 (25000/Abl x 10-4) and BAALC (450/Abl x 10-4) were arbitrary chosen after pre-analysis and comparison with our published data.Seven patients had FLT3-ITD mutation (17%), 18 (45%) patients had high WT1 expression levels and BAALC was overexpressed in 9 (23%) patients.
Results
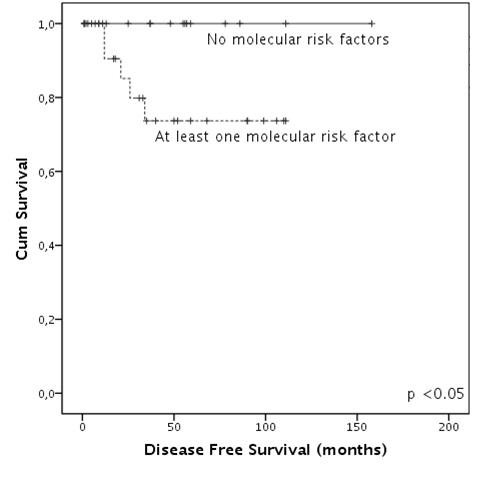
Sixty-three patients survived induction, all of them achieved CR. Among patients who had completed the whole therapeutic program at the time of the analysis 2 showed molecular persistence of disease and therefore received further therapy achieving molecular CR. Ten patients experienced disease relapse (15%).Pre-analysis showed that FLT3-ITD, not over-expressed WT1 levels, high BAALC levels but not the presence of BCR3 were associated with higher relapse risk. None of the molecular alterations reached statistical significance if taken alone. Therefore a molecular score including those 3 genes was built: 18 patients had no risk factor, 20 had one risk factor and 6 had two risk factors. Relapse risk was influenced only by the presence of at least one molecular risk factor (p<0.05).Disease free survival (DFS) duration was significantly higher in patients who had no molecular risk factor when compared to patients with at least one (3-years DFS of 100% and 85%, respectively, p<0.05). Overall survival (OS) analysis led to similar results: patients with no molecular risk factors had a very good outcome when compared to patients with at least one alteration (3-years OS of 100% and 79% respectively, p<0.03).Furthermore, we observed that both in DFS and OS analysis, the presence of two molecular factors leads to a worse outcome compared to the presence of only one alteration (p<0.05).
Conclusion
Even in a small cohort, we demonstrate how a simple molecular profile, including FLT3-ITD and levels of expression of BAALC and WT1 could identify a subset of standard APL patients at higher risk of relapse.Sub-stratification of patients could allow us to recognize patients who may take benefit from an intensified consolidation therapy, as it is currently applied to high risk patients, in order to reduce risk of relapse.
Session topic: E-poster
Keyword(s): Acute promyelocytic leukemia, Flt3-ITD, WT1
Type: Eposter Presentation
Background
Non high risk acute promyelocytic leukemia (APL) patients, as defined by WBC count at diagnosis, have nowadays a very good prognosis when treated with ATRA plus chemotherapy based protocols, with high rate of complete molecular remission after consolidation therapy. However, a small portion of patients (roughly 10%) will eventually relapse, despite the achievement of molecular CR.In the past years, some groups showed that relapse risk could be predicted by testing for some gene alterations, such as FLT3-ITD, or by break point cluster region (BCR) analysis. However, those results were not confirmed in prospective trials.
Aims
We recently showed that a simple gene-expression panel including WT1 and BAALC expression levels analysis has high prognostic value in MDS patients. The aim of the present study was to evaluate if the application of a similar panel to APL patients at diagnosis could be predictive of relapse risk and survival.
Methods
We retrospectively applied a simple 3 gene based molecular panel including WT1 and BAALC expression levels at diagnosis, FLT3-ITD mutational status (molecular profile), alongside with BCR analysis, to a cohort of de novo 66 non high-risk acute promyelocytic leukemia patients, treated in our institution between January 1st 2004 to June 30th 2015. All patients have been treated according to the Italian age-adapted AIDA protocol. Median age was 47 years (range 16-88), 17 patients were older than 60 years. Median follow up was 58 months.BCR analysis was available in all patients, 43 patients showed BCR1/2, whereas 23 patients had BCR3 (35%).Molecular profile was available in 44/66 patients (66%). Cut-off values for WT1 (25000/Abl x 10-4) and BAALC (450/Abl x 10-4) were arbitrary chosen after pre-analysis and comparison with our published data.Seven patients had FLT3-ITD mutation (17%), 18 (45%) patients had high WT1 expression levels and BAALC was overexpressed in 9 (23%) patients.
Results
Sixty-three patients survived induction, all of them achieved CR. Among patients who had completed the whole therapeutic program at the time of the analysis 2 showed molecular persistence of disease and therefore received further therapy achieving molecular CR. Ten patients experienced disease relapse (15%).Pre-analysis showed that FLT3-ITD, not over-expressed WT1 levels, high BAALC levels but not the presence of BCR3 were associated with higher relapse risk. None of the molecular alterations reached statistical significance if taken alone. Therefore a molecular score including those 3 genes was built: 18 patients had no risk factor, 20 had one risk factor and 6 had two risk factors. Relapse risk was influenced only by the presence of at least one molecular risk factor (p<0.05).Disease free survival (DFS) duration was significantly higher in patients who had no molecular risk factor when compared to patients with at least one (3-years DFS of 100% and 85%, respectively, p<0.05). Overall survival (OS) analysis led to similar results: patients with no molecular risk factors had a very good outcome when compared to patients with at least one alteration (3-years OS of 100% and 79% respectively, p<0.03).Furthermore, we observed that both in DFS and OS analysis, the presence of two molecular factors leads to a worse outcome compared to the presence of only one alteration (p<0.05).
Conclusion
Even in a small cohort, we demonstrate how a simple molecular profile, including FLT3-ITD and levels of expression of BAALC and WT1 could identify a subset of standard APL patients at higher risk of relapse.Sub-stratification of patients could allow us to recognize patients who may take benefit from an intensified consolidation therapy, as it is currently applied to high risk patients, in order to reduce risk of relapse.

Session topic: E-poster
Keyword(s): Acute promyelocytic leukemia, Flt3-ITD, WT1
Abstract: E921
Type: Eposter Presentation
Background
Non high risk acute promyelocytic leukemia (APL) patients, as defined by WBC count at diagnosis, have nowadays a very good prognosis when treated with ATRA plus chemotherapy based protocols, with high rate of complete molecular remission after consolidation therapy. However, a small portion of patients (roughly 10%) will eventually relapse, despite the achievement of molecular CR.In the past years, some groups showed that relapse risk could be predicted by testing for some gene alterations, such as FLT3-ITD, or by break point cluster region (BCR) analysis. However, those results were not confirmed in prospective trials.
Aims
We recently showed that a simple gene-expression panel including WT1 and BAALC expression levels analysis has high prognostic value in MDS patients. The aim of the present study was to evaluate if the application of a similar panel to APL patients at diagnosis could be predictive of relapse risk and survival.
Methods
We retrospectively applied a simple 3 gene based molecular panel including WT1 and BAALC expression levels at diagnosis, FLT3-ITD mutational status (molecular profile), alongside with BCR analysis, to a cohort of de novo 66 non high-risk acute promyelocytic leukemia patients, treated in our institution between January 1st 2004 to June 30th 2015. All patients have been treated according to the Italian age-adapted AIDA protocol. Median age was 47 years (range 16-88), 17 patients were older than 60 years. Median follow up was 58 months.BCR analysis was available in all patients, 43 patients showed BCR1/2, whereas 23 patients had BCR3 (35%).Molecular profile was available in 44/66 patients (66%). Cut-off values for WT1 (25000/Abl x 10-4) and BAALC (450/Abl x 10-4) were arbitrary chosen after pre-analysis and comparison with our published data.Seven patients had FLT3-ITD mutation (17%), 18 (45%) patients had high WT1 expression levels and BAALC was overexpressed in 9 (23%) patients.
Results
Sixty-three patients survived induction, all of them achieved CR. Among patients who had completed the whole therapeutic program at the time of the analysis 2 showed molecular persistence of disease and therefore received further therapy achieving molecular CR. Ten patients experienced disease relapse (15%).Pre-analysis showed that FLT3-ITD, not over-expressed WT1 levels, high BAALC levels but not the presence of BCR3 were associated with higher relapse risk. None of the molecular alterations reached statistical significance if taken alone. Therefore a molecular score including those 3 genes was built: 18 patients had no risk factor, 20 had one risk factor and 6 had two risk factors. Relapse risk was influenced only by the presence of at least one molecular risk factor (p<0.05).Disease free survival (DFS) duration was significantly higher in patients who had no molecular risk factor when compared to patients with at least one (3-years DFS of 100% and 85%, respectively, p<0.05). Overall survival (OS) analysis led to similar results: patients with no molecular risk factors had a very good outcome when compared to patients with at least one alteration (3-years OS of 100% and 79% respectively, p<0.03).Furthermore, we observed that both in DFS and OS analysis, the presence of two molecular factors leads to a worse outcome compared to the presence of only one alteration (p<0.05).
Conclusion
Even in a small cohort, we demonstrate how a simple molecular profile, including FLT3-ITD and levels of expression of BAALC and WT1 could identify a subset of standard APL patients at higher risk of relapse.Sub-stratification of patients could allow us to recognize patients who may take benefit from an intensified consolidation therapy, as it is currently applied to high risk patients, in order to reduce risk of relapse.

Session topic: E-poster
Keyword(s): Acute promyelocytic leukemia, Flt3-ITD, WT1
Type: Eposter Presentation
Background
Non high risk acute promyelocytic leukemia (APL) patients, as defined by WBC count at diagnosis, have nowadays a very good prognosis when treated with ATRA plus chemotherapy based protocols, with high rate of complete molecular remission after consolidation therapy. However, a small portion of patients (roughly 10%) will eventually relapse, despite the achievement of molecular CR.In the past years, some groups showed that relapse risk could be predicted by testing for some gene alterations, such as FLT3-ITD, or by break point cluster region (BCR) analysis. However, those results were not confirmed in prospective trials.
Aims
We recently showed that a simple gene-expression panel including WT1 and BAALC expression levels analysis has high prognostic value in MDS patients. The aim of the present study was to evaluate if the application of a similar panel to APL patients at diagnosis could be predictive of relapse risk and survival.
Methods
We retrospectively applied a simple 3 gene based molecular panel including WT1 and BAALC expression levels at diagnosis, FLT3-ITD mutational status (molecular profile), alongside with BCR analysis, to a cohort of de novo 66 non high-risk acute promyelocytic leukemia patients, treated in our institution between January 1st 2004 to June 30th 2015. All patients have been treated according to the Italian age-adapted AIDA protocol. Median age was 47 years (range 16-88), 17 patients were older than 60 years. Median follow up was 58 months.BCR analysis was available in all patients, 43 patients showed BCR1/2, whereas 23 patients had BCR3 (35%).Molecular profile was available in 44/66 patients (66%). Cut-off values for WT1 (25000/Abl x 10-4) and BAALC (450/Abl x 10-4) were arbitrary chosen after pre-analysis and comparison with our published data.Seven patients had FLT3-ITD mutation (17%), 18 (45%) patients had high WT1 expression levels and BAALC was overexpressed in 9 (23%) patients.
Results
Sixty-three patients survived induction, all of them achieved CR. Among patients who had completed the whole therapeutic program at the time of the analysis 2 showed molecular persistence of disease and therefore received further therapy achieving molecular CR. Ten patients experienced disease relapse (15%).Pre-analysis showed that FLT3-ITD, not over-expressed WT1 levels, high BAALC levels but not the presence of BCR3 were associated with higher relapse risk. None of the molecular alterations reached statistical significance if taken alone. Therefore a molecular score including those 3 genes was built: 18 patients had no risk factor, 20 had one risk factor and 6 had two risk factors. Relapse risk was influenced only by the presence of at least one molecular risk factor (p<0.05).Disease free survival (DFS) duration was significantly higher in patients who had no molecular risk factor when compared to patients with at least one (3-years DFS of 100% and 85%, respectively, p<0.05). Overall survival (OS) analysis led to similar results: patients with no molecular risk factors had a very good outcome when compared to patients with at least one alteration (3-years OS of 100% and 79% respectively, p<0.03).Furthermore, we observed that both in DFS and OS analysis, the presence of two molecular factors leads to a worse outcome compared to the presence of only one alteration (p<0.05).
Conclusion
Even in a small cohort, we demonstrate how a simple molecular profile, including FLT3-ITD and levels of expression of BAALC and WT1 could identify a subset of standard APL patients at higher risk of relapse.Sub-stratification of patients could allow us to recognize patients who may take benefit from an intensified consolidation therapy, as it is currently applied to high risk patients, in order to reduce risk of relapse.

Session topic: E-poster
Keyword(s): Acute promyelocytic leukemia, Flt3-ITD, WT1
{{ help_message }}
{{filter}}


