NEW DELETION OF JAK2 DETECTED BY SNP ARRAY IN ACUTE MYELOID LEUKEMIA: A ROLE IN OVERALL SURVIVAL
(Abstract release date: 05/19/16)
EHA Library. Guadagnuolo V. 06/09/16; 132459; E910

Dr. Viviana Guadagnuolo
Contributions
Contributions
Abstract
Abstract: E910
Type: Eposter Presentation
Background
In Acute Myeloid Leukemia (AML), a hematologic malignancy that originates in hematopoietic stem and myeloid progenitor cells, there is a strong need to develop new diagnostic and therapeutic options. Age and karyotype have been recognized as the most prominent prognostic factors in AML patients. Novel array-based technique-single-nucleotide polymorphism (SNP) microarray can detect cytogenetic lesions involve mostly structural alterations with losses or gains of chromosomic material. Those chromosomic abnormalities are predictive of response and are very important to define therapeutic strategies. SNP microarray can detect copy-neutral loss of heterozygosity (CN-LOH), which plays a role in oncogenesis by duplicating oncogenes, inhibiting tumor suppressors, and stimulating improper epigenetic programming.
Aims
Our objective is to evaluate the prognostic impact of these genetic alterations on clinical outcome.
Methods
We analyzed 310 AML patients (pts): 218 performed by SNP Array 6.0 (Affymetrix) and 92 performed by Cytoscan HD Array(Affymetrix) and also by Next Generation Sequencing (NGS)-WES HiSeq 2000 (Illumina). SNP Array data were analyzed by Nexus Copy Number™ v7.5 (BioDiscovery).
Results
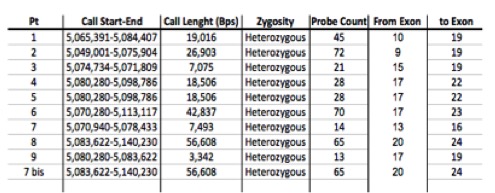
The median age of our cohort of pts was of 52 years and they had a median age at diagnosis of 51,3 years with a female/male ratio of 51% and 49%. Copy Number Alterations (CNAs) were detected in all patients affecting all the chromosomes, in particular our cohort of pts present a percentage of CNA, divided as follow: 42% of UPD (Uniparental Disomy), 19% of Copy Number (CN) gain, 39% of CN loss. By SNP array analysis we found that several genes were preferentially deleted, for example ADAM3A (62,2% pts), TP53 (15% pts), HRAS (8,6% pts), ETV6 (7,8% pts) and JAK2 (7% pts), while genes preferentially involved in amplifications are: FLT3 (42,5% pts), KIT (36,5% pts), KRAS (34,2% pts) and SIRPB1 (32,5% pts). We focused on a particular heterozygosis loss of JAK2, detected in 8% of pts, which goes from 5,049,001bp to 5,140,230p, involving the second tyrosine-kinase domain (figure 1). We showed that the group of patients which present this deletion had an overall survival rate better than the group with an amplification of the gene (p-value < 0,01). None CN loss in this region of JAK2 had been described in hematopoietic tissue before.
Conclusion
By SNP arrays we have identified CNAs involving important cancer genes in AML and we showed that a new deletion in JAK2 may play a role in the overall survival. Future prospectives will be to confirm and correlate other cancer genes aberrations and mutations with the prognosis of AML, in order to identify new biomarkers relevant for the disease.Acknowledgement: ELN, AIL, AIRC, PRIN, progetto Regione-Università 2010-12(L. Bolondi),FP7 NGS-PTL project.
Session topic: E-poster
Keyword(s): Acute myeloid leukemia, SNP
Type: Eposter Presentation
Background
In Acute Myeloid Leukemia (AML), a hematologic malignancy that originates in hematopoietic stem and myeloid progenitor cells, there is a strong need to develop new diagnostic and therapeutic options. Age and karyotype have been recognized as the most prominent prognostic factors in AML patients. Novel array-based technique-single-nucleotide polymorphism (SNP) microarray can detect cytogenetic lesions involve mostly structural alterations with losses or gains of chromosomic material. Those chromosomic abnormalities are predictive of response and are very important to define therapeutic strategies. SNP microarray can detect copy-neutral loss of heterozygosity (CN-LOH), which plays a role in oncogenesis by duplicating oncogenes, inhibiting tumor suppressors, and stimulating improper epigenetic programming.
Aims
Our objective is to evaluate the prognostic impact of these genetic alterations on clinical outcome.
Methods
We analyzed 310 AML patients (pts): 218 performed by SNP Array 6.0 (Affymetrix) and 92 performed by Cytoscan HD Array(Affymetrix) and also by Next Generation Sequencing (NGS)-WES HiSeq 2000 (Illumina). SNP Array data were analyzed by Nexus Copy Number™ v7.5 (BioDiscovery).
Results
The median age of our cohort of pts was of 52 years and they had a median age at diagnosis of 51,3 years with a female/male ratio of 51% and 49%. Copy Number Alterations (CNAs) were detected in all patients affecting all the chromosomes, in particular our cohort of pts present a percentage of CNA, divided as follow: 42% of UPD (Uniparental Disomy), 19% of Copy Number (CN) gain, 39% of CN loss. By SNP array analysis we found that several genes were preferentially deleted, for example ADAM3A (62,2% pts), TP53 (15% pts), HRAS (8,6% pts), ETV6 (7,8% pts) and JAK2 (7% pts), while genes preferentially involved in amplifications are: FLT3 (42,5% pts), KIT (36,5% pts), KRAS (34,2% pts) and SIRPB1 (32,5% pts). We focused on a particular heterozygosis loss of JAK2, detected in 8% of pts, which goes from 5,049,001bp to 5,140,230p, involving the second tyrosine-kinase domain (figure 1). We showed that the group of patients which present this deletion had an overall survival rate better than the group with an amplification of the gene (p-value < 0,01). None CN loss in this region of JAK2 had been described in hematopoietic tissue before.
Conclusion
By SNP arrays we have identified CNAs involving important cancer genes in AML and we showed that a new deletion in JAK2 may play a role in the overall survival. Future prospectives will be to confirm and correlate other cancer genes aberrations and mutations with the prognosis of AML, in order to identify new biomarkers relevant for the disease.Acknowledgement: ELN, AIL, AIRC, PRIN, progetto Regione-Università 2010-12(L. Bolondi),FP7 NGS-PTL project.

Session topic: E-poster
Keyword(s): Acute myeloid leukemia, SNP
Abstract: E910
Type: Eposter Presentation
Background
In Acute Myeloid Leukemia (AML), a hematologic malignancy that originates in hematopoietic stem and myeloid progenitor cells, there is a strong need to develop new diagnostic and therapeutic options. Age and karyotype have been recognized as the most prominent prognostic factors in AML patients. Novel array-based technique-single-nucleotide polymorphism (SNP) microarray can detect cytogenetic lesions involve mostly structural alterations with losses or gains of chromosomic material. Those chromosomic abnormalities are predictive of response and are very important to define therapeutic strategies. SNP microarray can detect copy-neutral loss of heterozygosity (CN-LOH), which plays a role in oncogenesis by duplicating oncogenes, inhibiting tumor suppressors, and stimulating improper epigenetic programming.
Aims
Our objective is to evaluate the prognostic impact of these genetic alterations on clinical outcome.
Methods
We analyzed 310 AML patients (pts): 218 performed by SNP Array 6.0 (Affymetrix) and 92 performed by Cytoscan HD Array(Affymetrix) and also by Next Generation Sequencing (NGS)-WES HiSeq 2000 (Illumina). SNP Array data were analyzed by Nexus Copy Number™ v7.5 (BioDiscovery).
Results
The median age of our cohort of pts was of 52 years and they had a median age at diagnosis of 51,3 years with a female/male ratio of 51% and 49%. Copy Number Alterations (CNAs) were detected in all patients affecting all the chromosomes, in particular our cohort of pts present a percentage of CNA, divided as follow: 42% of UPD (Uniparental Disomy), 19% of Copy Number (CN) gain, 39% of CN loss. By SNP array analysis we found that several genes were preferentially deleted, for example ADAM3A (62,2% pts), TP53 (15% pts), HRAS (8,6% pts), ETV6 (7,8% pts) and JAK2 (7% pts), while genes preferentially involved in amplifications are: FLT3 (42,5% pts), KIT (36,5% pts), KRAS (34,2% pts) and SIRPB1 (32,5% pts). We focused on a particular heterozygosis loss of JAK2, detected in 8% of pts, which goes from 5,049,001bp to 5,140,230p, involving the second tyrosine-kinase domain (figure 1). We showed that the group of patients which present this deletion had an overall survival rate better than the group with an amplification of the gene (p-value < 0,01). None CN loss in this region of JAK2 had been described in hematopoietic tissue before.
Conclusion
By SNP arrays we have identified CNAs involving important cancer genes in AML and we showed that a new deletion in JAK2 may play a role in the overall survival. Future prospectives will be to confirm and correlate other cancer genes aberrations and mutations with the prognosis of AML, in order to identify new biomarkers relevant for the disease.Acknowledgement: ELN, AIL, AIRC, PRIN, progetto Regione-Università 2010-12(L. Bolondi),FP7 NGS-PTL project.

Session topic: E-poster
Keyword(s): Acute myeloid leukemia, SNP
Type: Eposter Presentation
Background
In Acute Myeloid Leukemia (AML), a hematologic malignancy that originates in hematopoietic stem and myeloid progenitor cells, there is a strong need to develop new diagnostic and therapeutic options. Age and karyotype have been recognized as the most prominent prognostic factors in AML patients. Novel array-based technique-single-nucleotide polymorphism (SNP) microarray can detect cytogenetic lesions involve mostly structural alterations with losses or gains of chromosomic material. Those chromosomic abnormalities are predictive of response and are very important to define therapeutic strategies. SNP microarray can detect copy-neutral loss of heterozygosity (CN-LOH), which plays a role in oncogenesis by duplicating oncogenes, inhibiting tumor suppressors, and stimulating improper epigenetic programming.
Aims
Our objective is to evaluate the prognostic impact of these genetic alterations on clinical outcome.
Methods
We analyzed 310 AML patients (pts): 218 performed by SNP Array 6.0 (Affymetrix) and 92 performed by Cytoscan HD Array(Affymetrix) and also by Next Generation Sequencing (NGS)-WES HiSeq 2000 (Illumina). SNP Array data were analyzed by Nexus Copy Number™ v7.5 (BioDiscovery).
Results
The median age of our cohort of pts was of 52 years and they had a median age at diagnosis of 51,3 years with a female/male ratio of 51% and 49%. Copy Number Alterations (CNAs) were detected in all patients affecting all the chromosomes, in particular our cohort of pts present a percentage of CNA, divided as follow: 42% of UPD (Uniparental Disomy), 19% of Copy Number (CN) gain, 39% of CN loss. By SNP array analysis we found that several genes were preferentially deleted, for example ADAM3A (62,2% pts), TP53 (15% pts), HRAS (8,6% pts), ETV6 (7,8% pts) and JAK2 (7% pts), while genes preferentially involved in amplifications are: FLT3 (42,5% pts), KIT (36,5% pts), KRAS (34,2% pts) and SIRPB1 (32,5% pts). We focused on a particular heterozygosis loss of JAK2, detected in 8% of pts, which goes from 5,049,001bp to 5,140,230p, involving the second tyrosine-kinase domain (figure 1). We showed that the group of patients which present this deletion had an overall survival rate better than the group with an amplification of the gene (p-value < 0,01). None CN loss in this region of JAK2 had been described in hematopoietic tissue before.
Conclusion
By SNP arrays we have identified CNAs involving important cancer genes in AML and we showed that a new deletion in JAK2 may play a role in the overall survival. Future prospectives will be to confirm and correlate other cancer genes aberrations and mutations with the prognosis of AML, in order to identify new biomarkers relevant for the disease.Acknowledgement: ELN, AIL, AIRC, PRIN, progetto Regione-Università 2010-12(L. Bolondi),FP7 NGS-PTL project.

Session topic: E-poster
Keyword(s): Acute myeloid leukemia, SNP
{{ help_message }}
{{filter}}


