USE OF NGS MULTIGENIC PANEL IN MOLECULAR DIAGNOSTIC OF MYELOID MALIGNANCIES TO STRATIFY PATIENTS FOR PERSONALIZED THERAPIES
(Abstract release date: 05/19/16)
EHA Library. Franchini E. 06/09/16; 132454; E905

Dr. Eugenia Franchini
Contributions
Contributions
Abstract
Abstract: E905
Type: Eposter Presentation
Background
In the last years, next generation sequencing (NGS) technology resulted to be a new effective strategy in identifying genetic aberrations in myeloid neoplasms. Molecular mutation information became essential for biological subclassification, risk stratification and therapeutic decisions; the mutational status of several genes became important for understanding the complex interactions among different pathways in leukemogenesis.
Aims
Characterization of myeloid neoplasms using a multigenic panel of NGS sequencing in order to identify important alterations in a shorter time than traditional molecular methods and with a higher sensitivity. This will be helpful in a prematurely detection of small clones, important for monitoring disease progression and the inclusion in target therapy protocols.
Methods
We performed 15 runs with the TruSight Myeloid Panel of Illumina, for a total of 118 patients analyzed at diagnosis: 95 AML/sAML, 15 MPN, 3 CML, 3 MDS and 2 CMML. 25 patients (21,2%) had a normal karyotype, 25 (21,2%) presented one or two alterations, 26 (22%) had a complex karyotype, while for 42 patients (35,6%) no information about the karyotype was available. The panel is a next-generation sequencing platform to screen somatic variants in 54 genes relevant in myeloid diseases: 15 full genes (exons only) and oncogenic hotspots of 39 additional genes, for a total of 568 amplicons.
Results
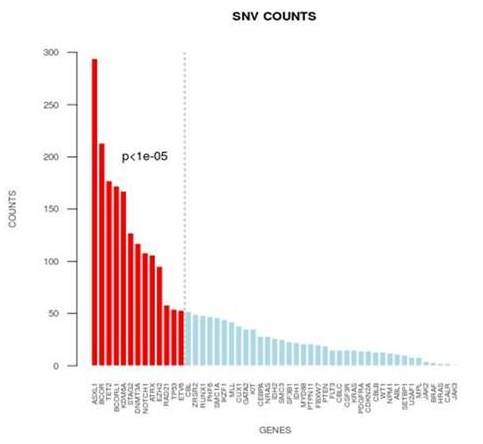
The output data were then analyzed with the Illumina’s software Variant Studio and the results were filtered by a coverage of minimum depth of 500 and allele frequency >3%. Variants already classified as SNP were removed. Only non-synonymous mutation were considered. The mean coverage was 3662 with a mean of 24 alterations per patient.57,5%, 3.9% and 38.6% of the alterations had a Variant Allele frequency (VAF) <10%, between 10-30% and >30%, respectively. Three patients resulted wild-type, while 10 patients carried only one mutation. The most mutated genes were ASXL1, BCOR, TET2, BCORL1, KDM6A, STAG2, DNMT3A, NOTCH1, ATRX, EZH2, RAD21, TP53, ETV6 (p<1e-05). We also detected 66 short deletions and 11 short insertions (max length 23 bp and 25 bp respectively).Then we proceeded with the validation of the mutations >10% of NPM1, DNMT3A, TP53, FLT3, JAK2, MPL, SETBP1, IDH1 and IDH2 with conventional molecular methods available in our laboratory (Sanger Sequencing and dPCR) and all the 145 mutations were confirmed. We also validated the alterations <10% of CEBPa, TP53, RUNX1, IDH1, IDH2, CALR and FLT3 with the Roche GS Junior 454 and we obtained >90% of concordance.Moreover 18 AML samples were also analyzed by WES and 97,5% of the mutations were confirmed.
Conclusion
These data suggest that a NGS multigenic panel is an effective strategy in myeloid neoplasms characterization and stratification in light of the development of novel personalized therapies (IDH-inhibitors: AG120 and AG221, MEK-inhibitors: GDC-0973, JAK-inibitors: Ruxolitinib and FLT3-inhibitors: Sorafenib, Midostaurin, AC220, ASP2215). Moreover, this approach is cheaper and time-saving and can also reveal alterations with a higher sensitivity than conventional methods. For this reason, we think that this approach could be strongly recommended for all new diagnosis/relapse myeloid neoplasm in order to obtain a more complete and premature characterization of the disease that will give advantages in term of therapeutic approach and OS of the patients.Akcnowledgments: work supported by ELN, AIL, AIRC, Progetto Regione-Università 2010-12 (L.Bolondi), FP7 NGS-PTL project, Illumina inc.
Session topic: E-poster
Keyword(s): Molecular markers, Myeloid malignancies, Targeted therapy
Type: Eposter Presentation
Background
In the last years, next generation sequencing (NGS) technology resulted to be a new effective strategy in identifying genetic aberrations in myeloid neoplasms. Molecular mutation information became essential for biological subclassification, risk stratification and therapeutic decisions; the mutational status of several genes became important for understanding the complex interactions among different pathways in leukemogenesis.
Aims
Characterization of myeloid neoplasms using a multigenic panel of NGS sequencing in order to identify important alterations in a shorter time than traditional molecular methods and with a higher sensitivity. This will be helpful in a prematurely detection of small clones, important for monitoring disease progression and the inclusion in target therapy protocols.
Methods
We performed 15 runs with the TruSight Myeloid Panel of Illumina, for a total of 118 patients analyzed at diagnosis: 95 AML/sAML, 15 MPN, 3 CML, 3 MDS and 2 CMML. 25 patients (21,2%) had a normal karyotype, 25 (21,2%) presented one or two alterations, 26 (22%) had a complex karyotype, while for 42 patients (35,6%) no information about the karyotype was available. The panel is a next-generation sequencing platform to screen somatic variants in 54 genes relevant in myeloid diseases: 15 full genes (exons only) and oncogenic hotspots of 39 additional genes, for a total of 568 amplicons.
Results
The output data were then analyzed with the Illumina’s software Variant Studio and the results were filtered by a coverage of minimum depth of 500 and allele frequency >3%. Variants already classified as SNP were removed. Only non-synonymous mutation were considered. The mean coverage was 3662 with a mean of 24 alterations per patient.57,5%, 3.9% and 38.6% of the alterations had a Variant Allele frequency (VAF) <10%, between 10-30% and >30%, respectively. Three patients resulted wild-type, while 10 patients carried only one mutation. The most mutated genes were ASXL1, BCOR, TET2, BCORL1, KDM6A, STAG2, DNMT3A, NOTCH1, ATRX, EZH2, RAD21, TP53, ETV6 (p<1e-05). We also detected 66 short deletions and 11 short insertions (max length 23 bp and 25 bp respectively).Then we proceeded with the validation of the mutations >10% of NPM1, DNMT3A, TP53, FLT3, JAK2, MPL, SETBP1, IDH1 and IDH2 with conventional molecular methods available in our laboratory (Sanger Sequencing and dPCR) and all the 145 mutations were confirmed. We also validated the alterations <10% of CEBPa, TP53, RUNX1, IDH1, IDH2, CALR and FLT3 with the Roche GS Junior 454 and we obtained >90% of concordance.Moreover 18 AML samples were also analyzed by WES and 97,5% of the mutations were confirmed.
Conclusion
These data suggest that a NGS multigenic panel is an effective strategy in myeloid neoplasms characterization and stratification in light of the development of novel personalized therapies (IDH-inhibitors: AG120 and AG221, MEK-inhibitors: GDC-0973, JAK-inibitors: Ruxolitinib and FLT3-inhibitors: Sorafenib, Midostaurin, AC220, ASP2215). Moreover, this approach is cheaper and time-saving and can also reveal alterations with a higher sensitivity than conventional methods. For this reason, we think that this approach could be strongly recommended for all new diagnosis/relapse myeloid neoplasm in order to obtain a more complete and premature characterization of the disease that will give advantages in term of therapeutic approach and OS of the patients.Akcnowledgments: work supported by ELN, AIL, AIRC, Progetto Regione-Università 2010-12 (L.Bolondi), FP7 NGS-PTL project, Illumina inc.

Session topic: E-poster
Keyword(s): Molecular markers, Myeloid malignancies, Targeted therapy
Abstract: E905
Type: Eposter Presentation
Background
In the last years, next generation sequencing (NGS) technology resulted to be a new effective strategy in identifying genetic aberrations in myeloid neoplasms. Molecular mutation information became essential for biological subclassification, risk stratification and therapeutic decisions; the mutational status of several genes became important for understanding the complex interactions among different pathways in leukemogenesis.
Aims
Characterization of myeloid neoplasms using a multigenic panel of NGS sequencing in order to identify important alterations in a shorter time than traditional molecular methods and with a higher sensitivity. This will be helpful in a prematurely detection of small clones, important for monitoring disease progression and the inclusion in target therapy protocols.
Methods
We performed 15 runs with the TruSight Myeloid Panel of Illumina, for a total of 118 patients analyzed at diagnosis: 95 AML/sAML, 15 MPN, 3 CML, 3 MDS and 2 CMML. 25 patients (21,2%) had a normal karyotype, 25 (21,2%) presented one or two alterations, 26 (22%) had a complex karyotype, while for 42 patients (35,6%) no information about the karyotype was available. The panel is a next-generation sequencing platform to screen somatic variants in 54 genes relevant in myeloid diseases: 15 full genes (exons only) and oncogenic hotspots of 39 additional genes, for a total of 568 amplicons.
Results
The output data were then analyzed with the Illumina’s software Variant Studio and the results were filtered by a coverage of minimum depth of 500 and allele frequency >3%. Variants already classified as SNP were removed. Only non-synonymous mutation were considered. The mean coverage was 3662 with a mean of 24 alterations per patient.57,5%, 3.9% and 38.6% of the alterations had a Variant Allele frequency (VAF) <10%, between 10-30% and >30%, respectively. Three patients resulted wild-type, while 10 patients carried only one mutation. The most mutated genes were ASXL1, BCOR, TET2, BCORL1, KDM6A, STAG2, DNMT3A, NOTCH1, ATRX, EZH2, RAD21, TP53, ETV6 (p<1e-05). We also detected 66 short deletions and 11 short insertions (max length 23 bp and 25 bp respectively).Then we proceeded with the validation of the mutations >10% of NPM1, DNMT3A, TP53, FLT3, JAK2, MPL, SETBP1, IDH1 and IDH2 with conventional molecular methods available in our laboratory (Sanger Sequencing and dPCR) and all the 145 mutations were confirmed. We also validated the alterations <10% of CEBPa, TP53, RUNX1, IDH1, IDH2, CALR and FLT3 with the Roche GS Junior 454 and we obtained >90% of concordance.Moreover 18 AML samples were also analyzed by WES and 97,5% of the mutations were confirmed.
Conclusion
These data suggest that a NGS multigenic panel is an effective strategy in myeloid neoplasms characterization and stratification in light of the development of novel personalized therapies (IDH-inhibitors: AG120 and AG221, MEK-inhibitors: GDC-0973, JAK-inibitors: Ruxolitinib and FLT3-inhibitors: Sorafenib, Midostaurin, AC220, ASP2215). Moreover, this approach is cheaper and time-saving and can also reveal alterations with a higher sensitivity than conventional methods. For this reason, we think that this approach could be strongly recommended for all new diagnosis/relapse myeloid neoplasm in order to obtain a more complete and premature characterization of the disease that will give advantages in term of therapeutic approach and OS of the patients.Akcnowledgments: work supported by ELN, AIL, AIRC, Progetto Regione-Università 2010-12 (L.Bolondi), FP7 NGS-PTL project, Illumina inc.

Session topic: E-poster
Keyword(s): Molecular markers, Myeloid malignancies, Targeted therapy
Type: Eposter Presentation
Background
In the last years, next generation sequencing (NGS) technology resulted to be a new effective strategy in identifying genetic aberrations in myeloid neoplasms. Molecular mutation information became essential for biological subclassification, risk stratification and therapeutic decisions; the mutational status of several genes became important for understanding the complex interactions among different pathways in leukemogenesis.
Aims
Characterization of myeloid neoplasms using a multigenic panel of NGS sequencing in order to identify important alterations in a shorter time than traditional molecular methods and with a higher sensitivity. This will be helpful in a prematurely detection of small clones, important for monitoring disease progression and the inclusion in target therapy protocols.
Methods
We performed 15 runs with the TruSight Myeloid Panel of Illumina, for a total of 118 patients analyzed at diagnosis: 95 AML/sAML, 15 MPN, 3 CML, 3 MDS and 2 CMML. 25 patients (21,2%) had a normal karyotype, 25 (21,2%) presented one or two alterations, 26 (22%) had a complex karyotype, while for 42 patients (35,6%) no information about the karyotype was available. The panel is a next-generation sequencing platform to screen somatic variants in 54 genes relevant in myeloid diseases: 15 full genes (exons only) and oncogenic hotspots of 39 additional genes, for a total of 568 amplicons.
Results
The output data were then analyzed with the Illumina’s software Variant Studio and the results were filtered by a coverage of minimum depth of 500 and allele frequency >3%. Variants already classified as SNP were removed. Only non-synonymous mutation were considered. The mean coverage was 3662 with a mean of 24 alterations per patient.57,5%, 3.9% and 38.6% of the alterations had a Variant Allele frequency (VAF) <10%, between 10-30% and >30%, respectively. Three patients resulted wild-type, while 10 patients carried only one mutation. The most mutated genes were ASXL1, BCOR, TET2, BCORL1, KDM6A, STAG2, DNMT3A, NOTCH1, ATRX, EZH2, RAD21, TP53, ETV6 (p<1e-05). We also detected 66 short deletions and 11 short insertions (max length 23 bp and 25 bp respectively).Then we proceeded with the validation of the mutations >10% of NPM1, DNMT3A, TP53, FLT3, JAK2, MPL, SETBP1, IDH1 and IDH2 with conventional molecular methods available in our laboratory (Sanger Sequencing and dPCR) and all the 145 mutations were confirmed. We also validated the alterations <10% of CEBPa, TP53, RUNX1, IDH1, IDH2, CALR and FLT3 with the Roche GS Junior 454 and we obtained >90% of concordance.Moreover 18 AML samples were also analyzed by WES and 97,5% of the mutations were confirmed.
Conclusion
These data suggest that a NGS multigenic panel is an effective strategy in myeloid neoplasms characterization and stratification in light of the development of novel personalized therapies (IDH-inhibitors: AG120 and AG221, MEK-inhibitors: GDC-0973, JAK-inibitors: Ruxolitinib and FLT3-inhibitors: Sorafenib, Midostaurin, AC220, ASP2215). Moreover, this approach is cheaper and time-saving and can also reveal alterations with a higher sensitivity than conventional methods. For this reason, we think that this approach could be strongly recommended for all new diagnosis/relapse myeloid neoplasm in order to obtain a more complete and premature characterization of the disease that will give advantages in term of therapeutic approach and OS of the patients.Akcnowledgments: work supported by ELN, AIL, AIRC, Progetto Regione-Università 2010-12 (L.Bolondi), FP7 NGS-PTL project, Illumina inc.

Session topic: E-poster
Keyword(s): Molecular markers, Myeloid malignancies, Targeted therapy
{{ help_message }}
{{filter}}


