MICRORNAS HSA-LET-7A AND MIR-142-3P IN T CELLS IN PATIENTS PRESENTING WITH ACUTE MYELOID LEUKAEMIA – A MEANS OF REPAIRING TUMOUR-INDUCED T CELL DEFECTS?
(Abstract release date: 05/19/16)
EHA Library. Petty R. 06/09/16; 132439; E890

Dr. Robert Petty
Contributions
Contributions
Abstract
Abstract: E890
Type: Eposter Presentation
Background
Our previous work has shown that the absolute number of T cells is increased in the peripheral blood of patients presenting with Acute Myeloid Leukaemia (AML). These T cells can form conjugates with tumour cells but subsequent immune synapse formation and recruitment of downstream signalling molecules is inhibited. We found an altered T cell gene expression profile indicative of aberrant TCR signalling and T cell activation patterns suggesting a blunted immune response. We now hypothesize that a means by which these alterations in T cell gene expression are effected is via modulation of microRNA (miRNA) expression.
Aims
To define miRNA deregulation in peripheral blood T cell populations in patients presenting with AML compared to healthy age-matched individuals.
Methods
In a pilot study, CD4 and CD8 T cells were isolated from stored peripheral blood samples taken from AML patients at diagnosis and pre-treatment (n=3) and age/sex matched healthy volunteers (n=3) by FACS (average purity 98%). All AML patients had intermediate cytogenetic risk. Total RNA was hybridised to custom small RNA microarrays (MD Anderson) containing probes for 18694 human miRNA and 30945 other non-coding RNA species. Duplicate normalized probe intensities were averaged and differentially expressed small RNA were determined by LIMMA (linear models for microarray data) taking a FDR p>0.05 and fold change of +/- 1.5. Unsupervised hierarchical clustering and principle component analysis (PCA) were performed in R studio V3.2.2. Supervised clustering was conducted in Cluster V3.0 and Treeview V1.1.6r4. Identification of targeted mRNA was performed using miRTarBase (www.mirtarbase.mbc.nctu.edu.tw/).
Results
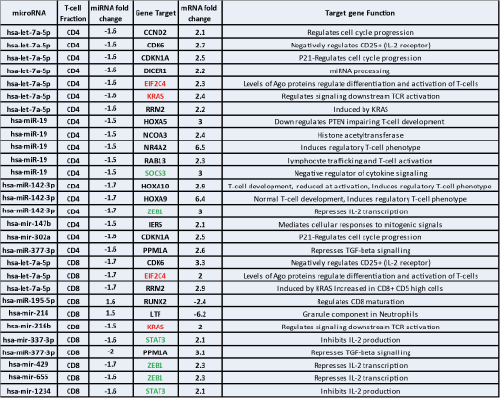
Hierarchical clustering and PCA of all samples, rather than differentiating samples by T cell population, differentiated AML from healthy controls illustrating that miRNA expression in T cells is altered by the presence of AML blasts. Supervised clustering demonstrated 39 miRNA were differentially expressed in AML CD4 T cells - 27 down-regulated, 12 up-regulated and 71 miRNA were differentially expressed in AML CD8 T cells - 41 down-regulated, 30 up-regulated. A number of these miRNA have already been shown to have a role in T cell proliferation and cellular migration. To validate this list, this dataset was correlated with our previously generated mRNA dataset (independent patient and control cohort) (see Figure). Crucially, this demonstrates 2 independent datasets showing coordinated deregulation of genes critical to T cell function. In both CD4 and CD8 cells, up-regulated genes are involved in T cell differentiation and activation (EIF2CA and KRAS) but also paradoxically in inhibition of cytokine production (STAT3 and ZEB1 in CD8 cells and SOCS3 and ZEB1 in CD4 cells) supporting our previous hypothesis of aberrant T cell activation in AML. Two candidate miRNAs whose expression may induce the observed T cell defects are hsa-let-7a-5p and miR-142-3p. Hsa-let-7a-5p is down-regulated in both CD4 and CD8 cells and has recognised functions in the regulation of T cell proliferation via CDK6 and MYC as well as in Th2 differentiation. miR-142-3p is down-regulated in CD4 cells and its reduced expression inhibits T cell proliferation via HOX10A.
Conclusion
These data support the hypothesis that T cell miRNA expression is altered by the presence of AML blasts. This microRNA dataset has been validated against an independent mRNA dataset. Potential miRNA of interest are hsa-let-7a-5p and miR-142-3p as they have fundamental roles in normal T cell function. These findings will now be examined in a larger cohort.
Session topic: E-poster
Keyword(s): Acute myeloid leukemia, Microarray analysis, T cell
Type: Eposter Presentation
Background
Our previous work has shown that the absolute number of T cells is increased in the peripheral blood of patients presenting with Acute Myeloid Leukaemia (AML). These T cells can form conjugates with tumour cells but subsequent immune synapse formation and recruitment of downstream signalling molecules is inhibited. We found an altered T cell gene expression profile indicative of aberrant TCR signalling and T cell activation patterns suggesting a blunted immune response. We now hypothesize that a means by which these alterations in T cell gene expression are effected is via modulation of microRNA (miRNA) expression.
Aims
To define miRNA deregulation in peripheral blood T cell populations in patients presenting with AML compared to healthy age-matched individuals.
Methods
In a pilot study, CD4 and CD8 T cells were isolated from stored peripheral blood samples taken from AML patients at diagnosis and pre-treatment (n=3) and age/sex matched healthy volunteers (n=3) by FACS (average purity 98%). All AML patients had intermediate cytogenetic risk. Total RNA was hybridised to custom small RNA microarrays (MD Anderson) containing probes for 18694 human miRNA and 30945 other non-coding RNA species. Duplicate normalized probe intensities were averaged and differentially expressed small RNA were determined by LIMMA (linear models for microarray data) taking a FDR p>0.05 and fold change of +/- 1.5. Unsupervised hierarchical clustering and principle component analysis (PCA) were performed in R studio V3.2.2. Supervised clustering was conducted in Cluster V3.0 and Treeview V1.1.6r4. Identification of targeted mRNA was performed using miRTarBase (www.mirtarbase.mbc.nctu.edu.tw/).
Results
Hierarchical clustering and PCA of all samples, rather than differentiating samples by T cell population, differentiated AML from healthy controls illustrating that miRNA expression in T cells is altered by the presence of AML blasts. Supervised clustering demonstrated 39 miRNA were differentially expressed in AML CD4 T cells - 27 down-regulated, 12 up-regulated and 71 miRNA were differentially expressed in AML CD8 T cells - 41 down-regulated, 30 up-regulated. A number of these miRNA have already been shown to have a role in T cell proliferation and cellular migration. To validate this list, this dataset was correlated with our previously generated mRNA dataset (independent patient and control cohort) (see Figure). Crucially, this demonstrates 2 independent datasets showing coordinated deregulation of genes critical to T cell function. In both CD4 and CD8 cells, up-regulated genes are involved in T cell differentiation and activation (EIF2CA and KRAS) but also paradoxically in inhibition of cytokine production (STAT3 and ZEB1 in CD8 cells and SOCS3 and ZEB1 in CD4 cells) supporting our previous hypothesis of aberrant T cell activation in AML. Two candidate miRNAs whose expression may induce the observed T cell defects are hsa-let-7a-5p and miR-142-3p. Hsa-let-7a-5p is down-regulated in both CD4 and CD8 cells and has recognised functions in the regulation of T cell proliferation via CDK6 and MYC as well as in Th2 differentiation. miR-142-3p is down-regulated in CD4 cells and its reduced expression inhibits T cell proliferation via HOX10A.
Conclusion
These data support the hypothesis that T cell miRNA expression is altered by the presence of AML blasts. This microRNA dataset has been validated against an independent mRNA dataset. Potential miRNA of interest are hsa-let-7a-5p and miR-142-3p as they have fundamental roles in normal T cell function. These findings will now be examined in a larger cohort.

Session topic: E-poster
Keyword(s): Acute myeloid leukemia, Microarray analysis, T cell
Abstract: E890
Type: Eposter Presentation
Background
Our previous work has shown that the absolute number of T cells is increased in the peripheral blood of patients presenting with Acute Myeloid Leukaemia (AML). These T cells can form conjugates with tumour cells but subsequent immune synapse formation and recruitment of downstream signalling molecules is inhibited. We found an altered T cell gene expression profile indicative of aberrant TCR signalling and T cell activation patterns suggesting a blunted immune response. We now hypothesize that a means by which these alterations in T cell gene expression are effected is via modulation of microRNA (miRNA) expression.
Aims
To define miRNA deregulation in peripheral blood T cell populations in patients presenting with AML compared to healthy age-matched individuals.
Methods
In a pilot study, CD4 and CD8 T cells were isolated from stored peripheral blood samples taken from AML patients at diagnosis and pre-treatment (n=3) and age/sex matched healthy volunteers (n=3) by FACS (average purity 98%). All AML patients had intermediate cytogenetic risk. Total RNA was hybridised to custom small RNA microarrays (MD Anderson) containing probes for 18694 human miRNA and 30945 other non-coding RNA species. Duplicate normalized probe intensities were averaged and differentially expressed small RNA were determined by LIMMA (linear models for microarray data) taking a FDR p>0.05 and fold change of +/- 1.5. Unsupervised hierarchical clustering and principle component analysis (PCA) were performed in R studio V3.2.2. Supervised clustering was conducted in Cluster V3.0 and Treeview V1.1.6r4. Identification of targeted mRNA was performed using miRTarBase (www.mirtarbase.mbc.nctu.edu.tw/).
Results
Hierarchical clustering and PCA of all samples, rather than differentiating samples by T cell population, differentiated AML from healthy controls illustrating that miRNA expression in T cells is altered by the presence of AML blasts. Supervised clustering demonstrated 39 miRNA were differentially expressed in AML CD4 T cells - 27 down-regulated, 12 up-regulated and 71 miRNA were differentially expressed in AML CD8 T cells - 41 down-regulated, 30 up-regulated. A number of these miRNA have already been shown to have a role in T cell proliferation and cellular migration. To validate this list, this dataset was correlated with our previously generated mRNA dataset (independent patient and control cohort) (see Figure). Crucially, this demonstrates 2 independent datasets showing coordinated deregulation of genes critical to T cell function. In both CD4 and CD8 cells, up-regulated genes are involved in T cell differentiation and activation (EIF2CA and KRAS) but also paradoxically in inhibition of cytokine production (STAT3 and ZEB1 in CD8 cells and SOCS3 and ZEB1 in CD4 cells) supporting our previous hypothesis of aberrant T cell activation in AML. Two candidate miRNAs whose expression may induce the observed T cell defects are hsa-let-7a-5p and miR-142-3p. Hsa-let-7a-5p is down-regulated in both CD4 and CD8 cells and has recognised functions in the regulation of T cell proliferation via CDK6 and MYC as well as in Th2 differentiation. miR-142-3p is down-regulated in CD4 cells and its reduced expression inhibits T cell proliferation via HOX10A.
Conclusion
These data support the hypothesis that T cell miRNA expression is altered by the presence of AML blasts. This microRNA dataset has been validated against an independent mRNA dataset. Potential miRNA of interest are hsa-let-7a-5p and miR-142-3p as they have fundamental roles in normal T cell function. These findings will now be examined in a larger cohort.

Session topic: E-poster
Keyword(s): Acute myeloid leukemia, Microarray analysis, T cell
Type: Eposter Presentation
Background
Our previous work has shown that the absolute number of T cells is increased in the peripheral blood of patients presenting with Acute Myeloid Leukaemia (AML). These T cells can form conjugates with tumour cells but subsequent immune synapse formation and recruitment of downstream signalling molecules is inhibited. We found an altered T cell gene expression profile indicative of aberrant TCR signalling and T cell activation patterns suggesting a blunted immune response. We now hypothesize that a means by which these alterations in T cell gene expression are effected is via modulation of microRNA (miRNA) expression.
Aims
To define miRNA deregulation in peripheral blood T cell populations in patients presenting with AML compared to healthy age-matched individuals.
Methods
In a pilot study, CD4 and CD8 T cells were isolated from stored peripheral blood samples taken from AML patients at diagnosis and pre-treatment (n=3) and age/sex matched healthy volunteers (n=3) by FACS (average purity 98%). All AML patients had intermediate cytogenetic risk. Total RNA was hybridised to custom small RNA microarrays (MD Anderson) containing probes for 18694 human miRNA and 30945 other non-coding RNA species. Duplicate normalized probe intensities were averaged and differentially expressed small RNA were determined by LIMMA (linear models for microarray data) taking a FDR p>0.05 and fold change of +/- 1.5. Unsupervised hierarchical clustering and principle component analysis (PCA) were performed in R studio V3.2.2. Supervised clustering was conducted in Cluster V3.0 and Treeview V1.1.6r4. Identification of targeted mRNA was performed using miRTarBase (www.mirtarbase.mbc.nctu.edu.tw/).
Results
Hierarchical clustering and PCA of all samples, rather than differentiating samples by T cell population, differentiated AML from healthy controls illustrating that miRNA expression in T cells is altered by the presence of AML blasts. Supervised clustering demonstrated 39 miRNA were differentially expressed in AML CD4 T cells - 27 down-regulated, 12 up-regulated and 71 miRNA were differentially expressed in AML CD8 T cells - 41 down-regulated, 30 up-regulated. A number of these miRNA have already been shown to have a role in T cell proliferation and cellular migration. To validate this list, this dataset was correlated with our previously generated mRNA dataset (independent patient and control cohort) (see Figure). Crucially, this demonstrates 2 independent datasets showing coordinated deregulation of genes critical to T cell function. In both CD4 and CD8 cells, up-regulated genes are involved in T cell differentiation and activation (EIF2CA and KRAS) but also paradoxically in inhibition of cytokine production (STAT3 and ZEB1 in CD8 cells and SOCS3 and ZEB1 in CD4 cells) supporting our previous hypothesis of aberrant T cell activation in AML. Two candidate miRNAs whose expression may induce the observed T cell defects are hsa-let-7a-5p and miR-142-3p. Hsa-let-7a-5p is down-regulated in both CD4 and CD8 cells and has recognised functions in the regulation of T cell proliferation via CDK6 and MYC as well as in Th2 differentiation. miR-142-3p is down-regulated in CD4 cells and its reduced expression inhibits T cell proliferation via HOX10A.
Conclusion
These data support the hypothesis that T cell miRNA expression is altered by the presence of AML blasts. This microRNA dataset has been validated against an independent mRNA dataset. Potential miRNA of interest are hsa-let-7a-5p and miR-142-3p as they have fundamental roles in normal T cell function. These findings will now be examined in a larger cohort.

Session topic: E-poster
Keyword(s): Acute myeloid leukemia, Microarray analysis, T cell
{{ help_message }}
{{filter}}


