IDENTIFICATION OF THE GENOMIC AND TRANSCRIPTOMIC CHANGES CORRELATED WITH THE EX VIVO RESISTANCE TO DAUNORUBICIN AND DOXORUBICIN IN PEDIATRIC ACUTE LEUKEMIAS
(Abstract release date: 05/19/16)
EHA Library. Laskowska J. 06/09/16; 132394; E845

Ms. Joanna Laskowska
Contributions
Contributions
Abstract
Abstract: E845
Type: Eposter Presentation
Background
Drug resistances in leukemias correlates with numerous genomic and transcriptomic changes, but the significant majority is still unidentified. Daunorubicin (DNR) and doxorubicin (DOX) are chemotherapeutics of the anthracycline family frequently used in acute leukemias (AL) treatment.
Aims
The objective of the experiment was to identify changes on the genome and transcriptome level and compare them with ex vivo resistance to DNR and DOX in pediatric patients with acute lymphoblastic (ALL) or myeloblastic (AML) leukemia.
Methods
In order to determine the ex vivo drug resistance profiles to DOX and DNR, MTT cytotoxicity assay was performed on mononuclear cells from 155 patients with ALL or AML. Gene expression profiles (51 patients with ALL, 16 with AML) were prepared on the basis of cRNA hybridization to oligonucleotide arrays of the human genome (Affymetrix). CGH array profiles, (34 patients with ALL, 12 with AML) were prepared on the basis of gDNA hybridization to oligonucleotide arrays of the human genome (Agilent). Data were analyzed using bioinformatics tools (Partek GS, GeneSpring, Feature Extraction, CytoGenomics) and data bases (UCSC Genome Browser, GeneCards, PantherDB). Validation of array results was performed by RT-qPCR with the use of UPL probes for 20 genes comparing to 4 reference genes. The relative expression was calculated by Pfaffl method of quantification in REST-2009 (Qiagen).
Results
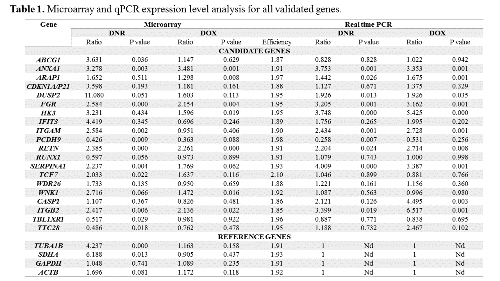
Ontological analysis of selected genes in transcriptomic profiles revealed, that hydrolases were mostly overexpressed in lymphoblasts resistant to DNR and DOX, but contrarily hydrolases showed differential expression level in AML cells resistant to DNR. Moreover, in both leukemic blasts resistant to DNR, chemokines were overexpressed. Additionally, in myeloblasts resistant to DNR, genes involved in transcription from RNA polymerase II promoter were downregulated, in ALL cells resistant to DOX – overexpressed, and in ALL blasts resistant to DNR genes showed differential expression. On the basis of achieved aCGH results, we singled out a couple of recurrent genomic changes identified among children with AL. Amp8p12-p11.21 is statistically significant rearrangement in resistant DNR profile, additionally del9p21.3 and amp22q11.22 are in resistant DOX profile. In blasts sensitive to DNR, we also identified del5q32-35.3, amp14q32.33, del15q11.1-11.2 and del21.11.2-p11.1. What is more, the results of relative expression analysis of candidate and reference genes are presented in Table 1.
Conclusion
The multidimensional analyses revealed, that resistance to DNR and DOX are the result of recurrent genomic and transcriptomic changes in leukemic blasts. We presume, that chromosome aberrations del9p21.3 and amp22q11.22 may associate with resistance to DOX, and amp8p12-p11.21 with lack of sensitivity to DNR and DOX.Additionally, basing on the Real-Time PCR results, we selected presumable target genes, such as ANXA1, ARAP1, FGR, HK3, SERP1, ITGAM, DUSP2, RETN, ITGB2 in DOX and IDA profiles. All of them were overexpressed. Interestingly, CASP1 was unique target gene in DOX resistant profile and PCDH9 in DNR resistant profile. Overexpression of CASP1 may influence on immune response in leukemia. Furthermore, the decreased expression level of cadherins (such as PCDH9) may have impact on the interactions of hematopoietic progenitors and bone marrow stromal cells, resulting breakdown of adhesive mechanisms and enhancement of cell proliferation.This study was supported by Grant from the National Science Centre No. DEC-2011/03/D/NZ5/05749.
Session topic: E-poster
Keyword(s): Anthracycline, CGH, Drug resistance, Gene expression profile
Type: Eposter Presentation
Background
Drug resistances in leukemias correlates with numerous genomic and transcriptomic changes, but the significant majority is still unidentified. Daunorubicin (DNR) and doxorubicin (DOX) are chemotherapeutics of the anthracycline family frequently used in acute leukemias (AL) treatment.
Aims
The objective of the experiment was to identify changes on the genome and transcriptome level and compare them with ex vivo resistance to DNR and DOX in pediatric patients with acute lymphoblastic (ALL) or myeloblastic (AML) leukemia.
Methods
In order to determine the ex vivo drug resistance profiles to DOX and DNR, MTT cytotoxicity assay was performed on mononuclear cells from 155 patients with ALL or AML. Gene expression profiles (51 patients with ALL, 16 with AML) were prepared on the basis of cRNA hybridization to oligonucleotide arrays of the human genome (Affymetrix). CGH array profiles, (34 patients with ALL, 12 with AML) were prepared on the basis of gDNA hybridization to oligonucleotide arrays of the human genome (Agilent). Data were analyzed using bioinformatics tools (Partek GS, GeneSpring, Feature Extraction, CytoGenomics) and data bases (UCSC Genome Browser, GeneCards, PantherDB). Validation of array results was performed by RT-qPCR with the use of UPL probes for 20 genes comparing to 4 reference genes. The relative expression was calculated by Pfaffl method of quantification in REST-2009 (Qiagen).
Results
Ontological analysis of selected genes in transcriptomic profiles revealed, that hydrolases were mostly overexpressed in lymphoblasts resistant to DNR and DOX, but contrarily hydrolases showed differential expression level in AML cells resistant to DNR. Moreover, in both leukemic blasts resistant to DNR, chemokines were overexpressed. Additionally, in myeloblasts resistant to DNR, genes involved in transcription from RNA polymerase II promoter were downregulated, in ALL cells resistant to DOX – overexpressed, and in ALL blasts resistant to DNR genes showed differential expression. On the basis of achieved aCGH results, we singled out a couple of recurrent genomic changes identified among children with AL. Amp8p12-p11.21 is statistically significant rearrangement in resistant DNR profile, additionally del9p21.3 and amp22q11.22 are in resistant DOX profile. In blasts sensitive to DNR, we also identified del5q32-35.3, amp14q32.33, del15q11.1-11.2 and del21.11.2-p11.1. What is more, the results of relative expression analysis of candidate and reference genes are presented in Table 1.
Conclusion
The multidimensional analyses revealed, that resistance to DNR and DOX are the result of recurrent genomic and transcriptomic changes in leukemic blasts. We presume, that chromosome aberrations del9p21.3 and amp22q11.22 may associate with resistance to DOX, and amp8p12-p11.21 with lack of sensitivity to DNR and DOX.Additionally, basing on the Real-Time PCR results, we selected presumable target genes, such as ANXA1, ARAP1, FGR, HK3, SERP1, ITGAM, DUSP2, RETN, ITGB2 in DOX and IDA profiles. All of them were overexpressed. Interestingly, CASP1 was unique target gene in DOX resistant profile and PCDH9 in DNR resistant profile. Overexpression of CASP1 may influence on immune response in leukemia. Furthermore, the decreased expression level of cadherins (such as PCDH9) may have impact on the interactions of hematopoietic progenitors and bone marrow stromal cells, resulting breakdown of adhesive mechanisms and enhancement of cell proliferation.This study was supported by Grant from the National Science Centre No. DEC-2011/03/D/NZ5/05749.

Session topic: E-poster
Keyword(s): Anthracycline, CGH, Drug resistance, Gene expression profile
Abstract: E845
Type: Eposter Presentation
Background
Drug resistances in leukemias correlates with numerous genomic and transcriptomic changes, but the significant majority is still unidentified. Daunorubicin (DNR) and doxorubicin (DOX) are chemotherapeutics of the anthracycline family frequently used in acute leukemias (AL) treatment.
Aims
The objective of the experiment was to identify changes on the genome and transcriptome level and compare them with ex vivo resistance to DNR and DOX in pediatric patients with acute lymphoblastic (ALL) or myeloblastic (AML) leukemia.
Methods
In order to determine the ex vivo drug resistance profiles to DOX and DNR, MTT cytotoxicity assay was performed on mononuclear cells from 155 patients with ALL or AML. Gene expression profiles (51 patients with ALL, 16 with AML) were prepared on the basis of cRNA hybridization to oligonucleotide arrays of the human genome (Affymetrix). CGH array profiles, (34 patients with ALL, 12 with AML) were prepared on the basis of gDNA hybridization to oligonucleotide arrays of the human genome (Agilent). Data were analyzed using bioinformatics tools (Partek GS, GeneSpring, Feature Extraction, CytoGenomics) and data bases (UCSC Genome Browser, GeneCards, PantherDB). Validation of array results was performed by RT-qPCR with the use of UPL probes for 20 genes comparing to 4 reference genes. The relative expression was calculated by Pfaffl method of quantification in REST-2009 (Qiagen).
Results
Ontological analysis of selected genes in transcriptomic profiles revealed, that hydrolases were mostly overexpressed in lymphoblasts resistant to DNR and DOX, but contrarily hydrolases showed differential expression level in AML cells resistant to DNR. Moreover, in both leukemic blasts resistant to DNR, chemokines were overexpressed. Additionally, in myeloblasts resistant to DNR, genes involved in transcription from RNA polymerase II promoter were downregulated, in ALL cells resistant to DOX – overexpressed, and in ALL blasts resistant to DNR genes showed differential expression. On the basis of achieved aCGH results, we singled out a couple of recurrent genomic changes identified among children with AL. Amp8p12-p11.21 is statistically significant rearrangement in resistant DNR profile, additionally del9p21.3 and amp22q11.22 are in resistant DOX profile. In blasts sensitive to DNR, we also identified del5q32-35.3, amp14q32.33, del15q11.1-11.2 and del21.11.2-p11.1. What is more, the results of relative expression analysis of candidate and reference genes are presented in Table 1.
Conclusion
The multidimensional analyses revealed, that resistance to DNR and DOX are the result of recurrent genomic and transcriptomic changes in leukemic blasts. We presume, that chromosome aberrations del9p21.3 and amp22q11.22 may associate with resistance to DOX, and amp8p12-p11.21 with lack of sensitivity to DNR and DOX.Additionally, basing on the Real-Time PCR results, we selected presumable target genes, such as ANXA1, ARAP1, FGR, HK3, SERP1, ITGAM, DUSP2, RETN, ITGB2 in DOX and IDA profiles. All of them were overexpressed. Interestingly, CASP1 was unique target gene in DOX resistant profile and PCDH9 in DNR resistant profile. Overexpression of CASP1 may influence on immune response in leukemia. Furthermore, the decreased expression level of cadherins (such as PCDH9) may have impact on the interactions of hematopoietic progenitors and bone marrow stromal cells, resulting breakdown of adhesive mechanisms and enhancement of cell proliferation.This study was supported by Grant from the National Science Centre No. DEC-2011/03/D/NZ5/05749.

Session topic: E-poster
Keyword(s): Anthracycline, CGH, Drug resistance, Gene expression profile
Type: Eposter Presentation
Background
Drug resistances in leukemias correlates with numerous genomic and transcriptomic changes, but the significant majority is still unidentified. Daunorubicin (DNR) and doxorubicin (DOX) are chemotherapeutics of the anthracycline family frequently used in acute leukemias (AL) treatment.
Aims
The objective of the experiment was to identify changes on the genome and transcriptome level and compare them with ex vivo resistance to DNR and DOX in pediatric patients with acute lymphoblastic (ALL) or myeloblastic (AML) leukemia.
Methods
In order to determine the ex vivo drug resistance profiles to DOX and DNR, MTT cytotoxicity assay was performed on mononuclear cells from 155 patients with ALL or AML. Gene expression profiles (51 patients with ALL, 16 with AML) were prepared on the basis of cRNA hybridization to oligonucleotide arrays of the human genome (Affymetrix). CGH array profiles, (34 patients with ALL, 12 with AML) were prepared on the basis of gDNA hybridization to oligonucleotide arrays of the human genome (Agilent). Data were analyzed using bioinformatics tools (Partek GS, GeneSpring, Feature Extraction, CytoGenomics) and data bases (UCSC Genome Browser, GeneCards, PantherDB). Validation of array results was performed by RT-qPCR with the use of UPL probes for 20 genes comparing to 4 reference genes. The relative expression was calculated by Pfaffl method of quantification in REST-2009 (Qiagen).
Results
Ontological analysis of selected genes in transcriptomic profiles revealed, that hydrolases were mostly overexpressed in lymphoblasts resistant to DNR and DOX, but contrarily hydrolases showed differential expression level in AML cells resistant to DNR. Moreover, in both leukemic blasts resistant to DNR, chemokines were overexpressed. Additionally, in myeloblasts resistant to DNR, genes involved in transcription from RNA polymerase II promoter were downregulated, in ALL cells resistant to DOX – overexpressed, and in ALL blasts resistant to DNR genes showed differential expression. On the basis of achieved aCGH results, we singled out a couple of recurrent genomic changes identified among children with AL. Amp8p12-p11.21 is statistically significant rearrangement in resistant DNR profile, additionally del9p21.3 and amp22q11.22 are in resistant DOX profile. In blasts sensitive to DNR, we also identified del5q32-35.3, amp14q32.33, del15q11.1-11.2 and del21.11.2-p11.1. What is more, the results of relative expression analysis of candidate and reference genes are presented in Table 1.
Conclusion
The multidimensional analyses revealed, that resistance to DNR and DOX are the result of recurrent genomic and transcriptomic changes in leukemic blasts. We presume, that chromosome aberrations del9p21.3 and amp22q11.22 may associate with resistance to DOX, and amp8p12-p11.21 with lack of sensitivity to DNR and DOX.Additionally, basing on the Real-Time PCR results, we selected presumable target genes, such as ANXA1, ARAP1, FGR, HK3, SERP1, ITGAM, DUSP2, RETN, ITGB2 in DOX and IDA profiles. All of them were overexpressed. Interestingly, CASP1 was unique target gene in DOX resistant profile and PCDH9 in DNR resistant profile. Overexpression of CASP1 may influence on immune response in leukemia. Furthermore, the decreased expression level of cadherins (such as PCDH9) may have impact on the interactions of hematopoietic progenitors and bone marrow stromal cells, resulting breakdown of adhesive mechanisms and enhancement of cell proliferation.This study was supported by Grant from the National Science Centre No. DEC-2011/03/D/NZ5/05749.

Session topic: E-poster
Keyword(s): Anthracycline, CGH, Drug resistance, Gene expression profile
{{ help_message }}
{{filter}}


