Cancer Research Center

Contributions
Type: Oral Presentation
Presentation during EHA20: From 13.06.2015 16:30 to 13.06.2015 16:45
Location: Room Lehar 1 + 2
Background
“ERG-related” childhood B cell precursor acute lymphoblastic leukemias (BCP-ALL) are characterized by a specific gene expression signature and by recurrent ERG intragenic deletion. ERG-related patients have a favourable outcome, despite a high incidence of IKFZ1 deletions. Nothing is known about non-coding (nc) RNA regulation in this leukemia subtype.
Aims
To investigate the main features of ERG-related patients by integrative genomic and gene expression analysis in an Italian cohort of B-others ALLs enrolled in the AIEOP ALL 2000 therapeutic protocol with emphasis on ncRNAs.
Methods
143 BCP-ALL patients lacking known genomic aberrations or a hyperdiploid karyotype (”B others”) were assigned to the ERG-related subtype according to their gene expression profile (GEP) and ERG and IKFZ1 aberrations were assessed by genomic analyses. A representative sub-cohort of “B others” including 8 ERG-related and 16 non-ERG-related samples were profiled by ncRNA microarray (Mirna1.0, Affymetrix) and validated by qRT-PCR.
Results
Non supervised clustering of 143 B-others patients identified a cluster of 35 samples (24.5%) in which 15 (45.5%) carried the ERG intragenic deletion. None of patients outside this “ERG-related” GEP cluster had ERG deletions. Similar to previous reports from BFM and EORTC childhood ALL protocols for the ERG deleted subgroup, our GEP defined ERG-related patients were characterized by more common IKZF1 deletions (35.3% compared with 26.2% in non-ERG related) and a favourable outcome, with a cumulative relapse incidence at 5 years of 6%±4.1 vs. 25.7%±4.3, p-value 0.03; (3/35 vs. 29/108 relapses).
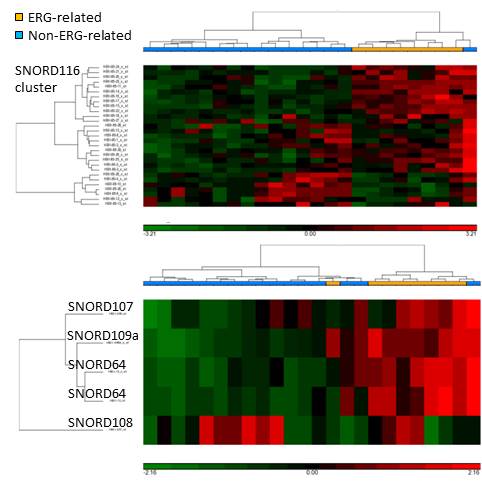
Analysis of ncRNAs revealed two interesting findings: Upregulation of the oncogenic miR-125b-2 cluster (hsa-miR-125b, -125b-2*, -99a, let-7c) and the related hsa-miR-100, hsa-miR-125a-3p, and a specific signature of snoRNAs tightly clustered within the ERG-related group. Strikingly the differentially expressed snoRNA probe sets (adjusted p-value <0.05) were located mainly in the Prader-Willi syndrome (PWS) locus at chromosome 15q11.2, a locus controlled by genomic imprinting. These snoRNAs are processed from a paternally expressed transcript that includes the PWS imprinting center upstream from SNURF/SNRPN. Specifically, there was up-regulation of SNORD109a, SNORD64, SNORD107 and 11 snoRNAs of the SNORD116 cluster (SNORD116-11, 14, 15, 16, 17, 20, 21, 22, 23, 24, 25) and downregulation of a subset of SNORD116 snoRNAs (SNORD116-4, 6, 10, 12, 13, 26) in ERG-related patients. Validation by qRT-PCR confirmed microarray findings.
Summary
ERG-related GEP defines a phenotypically unique subgroup in which only half carry ERG deletions, suggesting another underlying pathogenic mechanism beyond ERG intragenic deletion. We have identified a ncRNA signature specific of ERG-related BCP-ALL that can be used for differential diagnosis of these patients. While loss of imprinting associated with modified snoRNA expression was reported in acute promyelocytic leukemia, this is the first report of abnormal expression of snoRNAs in childhood BCP-ALL. The specific clustering of these snoRNAs within the imprinted PWS locus suggests a potential epigenetic mechanism underlying ERG-related ALLs.
Keyword(s): B cell acute lymphoblastic leukemia, Childhood, Molecular markers
Session topic: Biological pathway deregulation in B Cell Precursor ALL
Type: Oral Presentation
Presentation during EHA20: From 13.06.2015 16:30 to 13.06.2015 16:45
Location: Room Lehar 1 + 2
Background
“ERG-related” childhood B cell precursor acute lymphoblastic leukemias (BCP-ALL) are characterized by a specific gene expression signature and by recurrent ERG intragenic deletion. ERG-related patients have a favourable outcome, despite a high incidence of IKFZ1 deletions. Nothing is known about non-coding (nc) RNA regulation in this leukemia subtype.
Aims
To investigate the main features of ERG-related patients by integrative genomic and gene expression analysis in an Italian cohort of B-others ALLs enrolled in the AIEOP ALL 2000 therapeutic protocol with emphasis on ncRNAs.
Methods
143 BCP-ALL patients lacking known genomic aberrations or a hyperdiploid karyotype (”B others”) were assigned to the ERG-related subtype according to their gene expression profile (GEP) and ERG and IKFZ1 aberrations were assessed by genomic analyses. A representative sub-cohort of “B others” including 8 ERG-related and 16 non-ERG-related samples were profiled by ncRNA microarray (Mirna1.0, Affymetrix) and validated by qRT-PCR.
Results
Non supervised clustering of 143 B-others patients identified a cluster of 35 samples (24.5%) in which 15 (45.5%) carried the ERG intragenic deletion. None of patients outside this “ERG-related” GEP cluster had ERG deletions. Similar to previous reports from BFM and EORTC childhood ALL protocols for the ERG deleted subgroup, our GEP defined ERG-related patients were characterized by more common IKZF1 deletions (35.3% compared with 26.2% in non-ERG related) and a favourable outcome, with a cumulative relapse incidence at 5 years of 6%±4.1 vs. 25.7%±4.3, p-value 0.03; (3/35 vs. 29/108 relapses).
Analysis of ncRNAs revealed two interesting findings: Upregulation of the oncogenic miR-125b-2 cluster (hsa-miR-125b, -125b-2*, -99a, let-7c) and the related hsa-miR-100, hsa-miR-125a-3p, and a specific signature of snoRNAs tightly clustered within the ERG-related group. Strikingly the differentially expressed snoRNA probe sets (adjusted p-value <0.05) were located mainly in the Prader-Willi syndrome (PWS) locus at chromosome 15q11.2, a locus controlled by genomic imprinting. These snoRNAs are processed from a paternally expressed transcript that includes the PWS imprinting center upstream from SNURF/SNRPN. Specifically, there was up-regulation of SNORD109a, SNORD64, SNORD107 and 11 snoRNAs of the SNORD116 cluster (SNORD116-11, 14, 15, 16, 17, 20, 21, 22, 23, 24, 25) and downregulation of a subset of SNORD116 snoRNAs (SNORD116-4, 6, 10, 12, 13, 26) in ERG-related patients. Validation by qRT-PCR confirmed microarray findings.
Summary
ERG-related GEP defines a phenotypically unique subgroup in which only half carry ERG deletions, suggesting another underlying pathogenic mechanism beyond ERG intragenic deletion. We have identified a ncRNA signature specific of ERG-related BCP-ALL that can be used for differential diagnosis of these patients. While loss of imprinting associated with modified snoRNA expression was reported in acute promyelocytic leukemia, this is the first report of abnormal expression of snoRNAs in childhood BCP-ALL. The specific clustering of these snoRNAs within the imprinted PWS locus suggests a potential epigenetic mechanism underlying ERG-related ALLs.
Keyword(s): B cell acute lymphoblastic leukemia, Childhood, Molecular markers

Session topic: Biological pathway deregulation in B Cell Precursor ALL


