GENOMIC DNA BREAKPOINTS IN MLL AND TRANSLOCATION PARTNER GENES IN A LARGE COHORT OF INFANTS WITH ACUTE LEUKEMIA
(Abstract release date: 05/21/15)
EHA Library. Tsaur G. 06/13/15; 103220; S439
Disclosure(s): Regional Children's Hospital #1, Research Institute of Medical Cell technologiesPediatric Oncology and Hematology Center

Dr. Grigory Tsaur
Contributions
Contributions
Abstract
Abstract: S439
Type: Oral Presentation
Presentation during EHA20: From 13.06.2015 12:15 to 13.06.2015 12:30
Location: Room C1
Background
Acute leukemia (AL) in infants is characterized by high incidence of MLL gene rearrangements.
Aims
To evaluate the relation between genomic DNA breakpoints in MLL and translocation partner genes (TPGs) and clinical parameters of infant AL.
Methods
87 infants (32 boys (37%) and 55 girls (63%), median age 4.9 mo) with MLL-rearranged acute lymphoblastic leukemia (ALL) (n=63), acute myeloid leukemia (AML) (n=22) and mixed phenotype acute leukemia (MPAL) (n=2) were included in the current study. Genomic DNA breakpoint detection in MLL gene and translocation partner genes (TPGs) was performed by long-distance inverse PCR (LDI-PCR). Exon-intron numbering of MLL gene was done according to I. Nilson et al, 1996.
Results
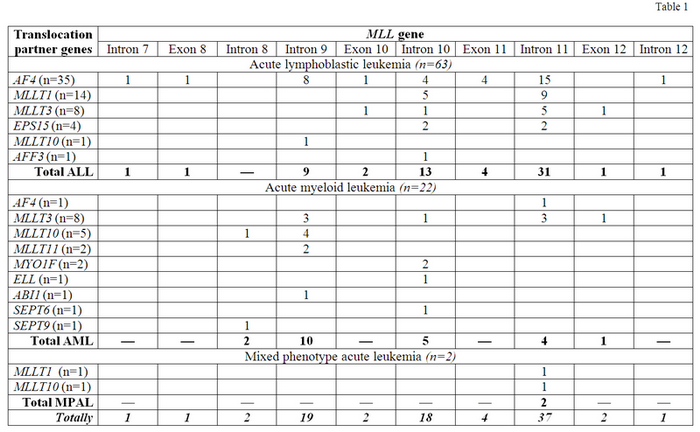
Majority of ALL cases was characterized by presence of MLL-AF4 fusion gene (FG) (n=35;55%), less frequently MLL-MLLT1 (n=12;22%), MLL-MLLT3 (n=8;13%) and others were found (Table 1). The most common breakpoint location within MLL gene in ALL patients was intron 11, detected in 31 cases (49%), less frequently breakpoints in intron 10 (n=13;21%) and intron 9 (n=9;14%) were found. The highest variability of MLL breakpoints was found in MLL-AF4-positive patients: only 15 of 35 (43%) had breakpoints in intron 11. The most stable pattern of MLL genomic DNA breakpoints was observed in MLL-MLLT1-positive patients: 9 of 14 (64%) had breakpoints in intron 11. In AML patients the most prevalent FG was MLL-MLLT3 (n=8;36%). The remaining ones are listed in Table 1. The most frequent breakpoint location was intron 9 (n=10;45%), less often they were found in intron 10 (n=5;23%) and 11 (n=4;18%). The most stable pattern was revealed for MLL-MLLT10 FG: MLL breakpoints in 4 of 5 (80%) cases were found in intron 9 (Table 1). In TPGs the most frequent breakpoint locations were as follows: in AF4 – intron 3 (n=25;69%) and intron 4 (n=7;19%); in MLLT1 – non-coding region between ACER1 and MLLT1 (n=9;60%) and intron 1 (n=5;33%); in MLLT3 – intron 5 (n=13;81%); in MLLT10 – intron 8 and intron 9 (n=2;33% each); in EPS15 – intron 1 (n=3;75%). The pattern of breakpoints locations in TPGs was similar in ALL, AML and MPAL cases. Distribution of DNA breakpoints in MLL gene was similar in boys and girls and did not depend on type of TPG. ALL patients who had breakpoints in intron 11 were significantly younger (median 3.0 mo, range 0.03-11.6) than all others (median 5.6 mo, range 0.7-11.9) (p=0.025) and than patients with MLL breakpoints in intron 9 (median 6.6 mo, range 3.1-11.9) (p=0.017). For AML cases we did not find any relation between age and breakpoints locations. We estimated prognostic significance of MLL breakpoint locations in 46 cases of infant ALL homogenously treated by MLL-Baby protocol. 5-year EFS was significantly lower in patients with breakpoint in intron 11 (n=29) in comparison to patients with breakpoints localized from intron 7 to exon 11 (n=17) (0.16±0.07 vs 0.38±0.14 p=0.035). While cumulative incidence of relapse was remarkably higher in the first group of patients (0.80±0.33 vs 0.56±0.20 p=0.020). Median follow-up time was 36 months. Although in Cox regression model including breakpoint position together with age, immunophenotype, initial WBC count, initial CNS involvement, type of MLL rearrangement, absolute blast number at day 8 of dexamethasone profase, minimal residual disease (MRD) at time point 4 (TP4) of MLL-Baby protocol, the only significant covariate was the presence of MRD at TP4 (HR 3.160, 95% CI 1.159-17.815, p=0.021).
Summary
Keyword(s): Acute lymphoblastic leukemia, Acute myeloid leukemia, Infant, MLL
Session topic: Translational studies in ALL
Type: Oral Presentation
Presentation during EHA20: From 13.06.2015 12:15 to 13.06.2015 12:30
Location: Room C1
Background
Acute leukemia (AL) in infants is characterized by high incidence of MLL gene rearrangements.
Aims
To evaluate the relation between genomic DNA breakpoints in MLL and translocation partner genes (TPGs) and clinical parameters of infant AL.
Methods
87 infants (32 boys (37%) and 55 girls (63%), median age 4.9 mo) with MLL-rearranged acute lymphoblastic leukemia (ALL) (n=63), acute myeloid leukemia (AML) (n=22) and mixed phenotype acute leukemia (MPAL) (n=2) were included in the current study. Genomic DNA breakpoint detection in MLL gene and translocation partner genes (TPGs) was performed by long-distance inverse PCR (LDI-PCR). Exon-intron numbering of MLL gene was done according to I. Nilson et al, 1996.
Results
Majority of ALL cases was characterized by presence of MLL-AF4 fusion gene (FG) (n=35;55%), less frequently MLL-MLLT1 (n=12;22%), MLL-MLLT3 (n=8;13%) and others were found (Table 1). The most common breakpoint location within MLL gene in ALL patients was intron 11, detected in 31 cases (49%), less frequently breakpoints in intron 10 (n=13;21%) and intron 9 (n=9;14%) were found. The highest variability of MLL breakpoints was found in MLL-AF4-positive patients: only 15 of 35 (43%) had breakpoints in intron 11. The most stable pattern of MLL genomic DNA breakpoints was observed in MLL-MLLT1-positive patients: 9 of 14 (64%) had breakpoints in intron 11. In AML patients the most prevalent FG was MLL-MLLT3 (n=8;36%). The remaining ones are listed in Table 1. The most frequent breakpoint location was intron 9 (n=10;45%), less often they were found in intron 10 (n=5;23%) and 11 (n=4;18%). The most stable pattern was revealed for MLL-MLLT10 FG: MLL breakpoints in 4 of 5 (80%) cases were found in intron 9 (Table 1). In TPGs the most frequent breakpoint locations were as follows: in AF4 – intron 3 (n=25;69%) and intron 4 (n=7;19%); in MLLT1 – non-coding region between ACER1 and MLLT1 (n=9;60%) and intron 1 (n=5;33%); in MLLT3 – intron 5 (n=13;81%); in MLLT10 – intron 8 and intron 9 (n=2;33% each); in EPS15 – intron 1 (n=3;75%). The pattern of breakpoints locations in TPGs was similar in ALL, AML and MPAL cases. Distribution of DNA breakpoints in MLL gene was similar in boys and girls and did not depend on type of TPG. ALL patients who had breakpoints in intron 11 were significantly younger (median 3.0 mo, range 0.03-11.6) than all others (median 5.6 mo, range 0.7-11.9) (p=0.025) and than patients with MLL breakpoints in intron 9 (median 6.6 mo, range 3.1-11.9) (p=0.017). For AML cases we did not find any relation between age and breakpoints locations. We estimated prognostic significance of MLL breakpoint locations in 46 cases of infant ALL homogenously treated by MLL-Baby protocol. 5-year EFS was significantly lower in patients with breakpoint in intron 11 (n=29) in comparison to patients with breakpoints localized from intron 7 to exon 11 (n=17) (0.16±0.07 vs 0.38±0.14 p=0.035). While cumulative incidence of relapse was remarkably higher in the first group of patients (0.80±0.33 vs 0.56±0.20 p=0.020). Median follow-up time was 36 months. Although in Cox regression model including breakpoint position together with age, immunophenotype, initial WBC count, initial CNS involvement, type of MLL rearrangement, absolute blast number at day 8 of dexamethasone profase, minimal residual disease (MRD) at time point 4 (TP4) of MLL-Baby protocol, the only significant covariate was the presence of MRD at TP4 (HR 3.160, 95% CI 1.159-17.815, p=0.021).
Summary
Our data provide additional information of molecular genetic features of MLL-rearranged infant AL.
Keyword(s): Acute lymphoblastic leukemia, Acute myeloid leukemia, Infant, MLL

Session topic: Translational studies in ALL
Abstract: S439
Type: Oral Presentation
Presentation during EHA20: From 13.06.2015 12:15 to 13.06.2015 12:30
Location: Room C1
Background
Acute leukemia (AL) in infants is characterized by high incidence of MLL gene rearrangements.
Aims
To evaluate the relation between genomic DNA breakpoints in MLL and translocation partner genes (TPGs) and clinical parameters of infant AL.
Methods
87 infants (32 boys (37%) and 55 girls (63%), median age 4.9 mo) with MLL-rearranged acute lymphoblastic leukemia (ALL) (n=63), acute myeloid leukemia (AML) (n=22) and mixed phenotype acute leukemia (MPAL) (n=2) were included in the current study. Genomic DNA breakpoint detection in MLL gene and translocation partner genes (TPGs) was performed by long-distance inverse PCR (LDI-PCR). Exon-intron numbering of MLL gene was done according to I. Nilson et al, 1996.
Results
Majority of ALL cases was characterized by presence of MLL-AF4 fusion gene (FG) (n=35;55%), less frequently MLL-MLLT1 (n=12;22%), MLL-MLLT3 (n=8;13%) and others were found (Table 1). The most common breakpoint location within MLL gene in ALL patients was intron 11, detected in 31 cases (49%), less frequently breakpoints in intron 10 (n=13;21%) and intron 9 (n=9;14%) were found. The highest variability of MLL breakpoints was found in MLL-AF4-positive patients: only 15 of 35 (43%) had breakpoints in intron 11. The most stable pattern of MLL genomic DNA breakpoints was observed in MLL-MLLT1-positive patients: 9 of 14 (64%) had breakpoints in intron 11. In AML patients the most prevalent FG was MLL-MLLT3 (n=8;36%). The remaining ones are listed in Table 1. The most frequent breakpoint location was intron 9 (n=10;45%), less often they were found in intron 10 (n=5;23%) and 11 (n=4;18%). The most stable pattern was revealed for MLL-MLLT10 FG: MLL breakpoints in 4 of 5 (80%) cases were found in intron 9 (Table 1). In TPGs the most frequent breakpoint locations were as follows: in AF4 – intron 3 (n=25;69%) and intron 4 (n=7;19%); in MLLT1 – non-coding region between ACER1 and MLLT1 (n=9;60%) and intron 1 (n=5;33%); in MLLT3 – intron 5 (n=13;81%); in MLLT10 – intron 8 and intron 9 (n=2;33% each); in EPS15 – intron 1 (n=3;75%). The pattern of breakpoints locations in TPGs was similar in ALL, AML and MPAL cases. Distribution of DNA breakpoints in MLL gene was similar in boys and girls and did not depend on type of TPG. ALL patients who had breakpoints in intron 11 were significantly younger (median 3.0 mo, range 0.03-11.6) than all others (median 5.6 mo, range 0.7-11.9) (p=0.025) and than patients with MLL breakpoints in intron 9 (median 6.6 mo, range 3.1-11.9) (p=0.017). For AML cases we did not find any relation between age and breakpoints locations. We estimated prognostic significance of MLL breakpoint locations in 46 cases of infant ALL homogenously treated by MLL-Baby protocol. 5-year EFS was significantly lower in patients with breakpoint in intron 11 (n=29) in comparison to patients with breakpoints localized from intron 7 to exon 11 (n=17) (0.16±0.07 vs 0.38±0.14 p=0.035). While cumulative incidence of relapse was remarkably higher in the first group of patients (0.80±0.33 vs 0.56±0.20 p=0.020). Median follow-up time was 36 months. Although in Cox regression model including breakpoint position together with age, immunophenotype, initial WBC count, initial CNS involvement, type of MLL rearrangement, absolute blast number at day 8 of dexamethasone profase, minimal residual disease (MRD) at time point 4 (TP4) of MLL-Baby protocol, the only significant covariate was the presence of MRD at TP4 (HR 3.160, 95% CI 1.159-17.815, p=0.021).
Summary
Keyword(s): Acute lymphoblastic leukemia, Acute myeloid leukemia, Infant, MLL

Session topic: Translational studies in ALL
Type: Oral Presentation
Presentation during EHA20: From 13.06.2015 12:15 to 13.06.2015 12:30
Location: Room C1
Background
Acute leukemia (AL) in infants is characterized by high incidence of MLL gene rearrangements.
Aims
To evaluate the relation between genomic DNA breakpoints in MLL and translocation partner genes (TPGs) and clinical parameters of infant AL.
Methods
87 infants (32 boys (37%) and 55 girls (63%), median age 4.9 mo) with MLL-rearranged acute lymphoblastic leukemia (ALL) (n=63), acute myeloid leukemia (AML) (n=22) and mixed phenotype acute leukemia (MPAL) (n=2) were included in the current study. Genomic DNA breakpoint detection in MLL gene and translocation partner genes (TPGs) was performed by long-distance inverse PCR (LDI-PCR). Exon-intron numbering of MLL gene was done according to I. Nilson et al, 1996.
Results
Majority of ALL cases was characterized by presence of MLL-AF4 fusion gene (FG) (n=35;55%), less frequently MLL-MLLT1 (n=12;22%), MLL-MLLT3 (n=8;13%) and others were found (Table 1). The most common breakpoint location within MLL gene in ALL patients was intron 11, detected in 31 cases (49%), less frequently breakpoints in intron 10 (n=13;21%) and intron 9 (n=9;14%) were found. The highest variability of MLL breakpoints was found in MLL-AF4-positive patients: only 15 of 35 (43%) had breakpoints in intron 11. The most stable pattern of MLL genomic DNA breakpoints was observed in MLL-MLLT1-positive patients: 9 of 14 (64%) had breakpoints in intron 11. In AML patients the most prevalent FG was MLL-MLLT3 (n=8;36%). The remaining ones are listed in Table 1. The most frequent breakpoint location was intron 9 (n=10;45%), less often they were found in intron 10 (n=5;23%) and 11 (n=4;18%). The most stable pattern was revealed for MLL-MLLT10 FG: MLL breakpoints in 4 of 5 (80%) cases were found in intron 9 (Table 1). In TPGs the most frequent breakpoint locations were as follows: in AF4 – intron 3 (n=25;69%) and intron 4 (n=7;19%); in MLLT1 – non-coding region between ACER1 and MLLT1 (n=9;60%) and intron 1 (n=5;33%); in MLLT3 – intron 5 (n=13;81%); in MLLT10 – intron 8 and intron 9 (n=2;33% each); in EPS15 – intron 1 (n=3;75%). The pattern of breakpoints locations in TPGs was similar in ALL, AML and MPAL cases. Distribution of DNA breakpoints in MLL gene was similar in boys and girls and did not depend on type of TPG. ALL patients who had breakpoints in intron 11 were significantly younger (median 3.0 mo, range 0.03-11.6) than all others (median 5.6 mo, range 0.7-11.9) (p=0.025) and than patients with MLL breakpoints in intron 9 (median 6.6 mo, range 3.1-11.9) (p=0.017). For AML cases we did not find any relation between age and breakpoints locations. We estimated prognostic significance of MLL breakpoint locations in 46 cases of infant ALL homogenously treated by MLL-Baby protocol. 5-year EFS was significantly lower in patients with breakpoint in intron 11 (n=29) in comparison to patients with breakpoints localized from intron 7 to exon 11 (n=17) (0.16±0.07 vs 0.38±0.14 p=0.035). While cumulative incidence of relapse was remarkably higher in the first group of patients (0.80±0.33 vs 0.56±0.20 p=0.020). Median follow-up time was 36 months. Although in Cox regression model including breakpoint position together with age, immunophenotype, initial WBC count, initial CNS involvement, type of MLL rearrangement, absolute blast number at day 8 of dexamethasone profase, minimal residual disease (MRD) at time point 4 (TP4) of MLL-Baby protocol, the only significant covariate was the presence of MRD at TP4 (HR 3.160, 95% CI 1.159-17.815, p=0.021).
Summary
Our data provide additional information of molecular genetic features of MLL-rearranged infant AL.
Keyword(s): Acute lymphoblastic leukemia, Acute myeloid leukemia, Infant, MLL

Session topic: Translational studies in ALL
{{ help_message }}
{{filter}}


