LLR Molecular Haematology Unit, Nuffield Division of Clinical Laboratory Sciences, Radcliffe Department of Medicine

Contributions
Type: Oral Presentation + travel grant
Presentation during EHA20: From 13.06.2015 12:00 to 13.06.2015 12:15
Location: Room Stolz 1
Background
The CRISPR/Cas9 genome editing system has recently emerged as a powerful tool for genome engineering, and has been used for gene correction in cultured cells from patients with monogenic hereditary defects. We reasoned that this technology could be employed to correct acquired gene mutations in human cancer cells.
Recurrent somatic mutations of the epigenetic modifier ASXL1 are common in myeloid malignancies, including myelodysplastic syndromes and chronic myeloid leukemia (CML). Loss-of-function ASXL1 mutations are strongly associated with a poor prognosis in these disorders. We studied the CML line KBM5 which lacks wild-type ASXL1 protein expression, due to a homozygous nonsense mutation (G710X) in the ASXL1 gene.
Aims
We used CRISPR/Cas9-mediated homology directed repair (HDR) to correct the ASXL1 mutation in KBM5 cells and performed functional studies to determine whether the wild-type function of ASXL1 was restored.
Methods
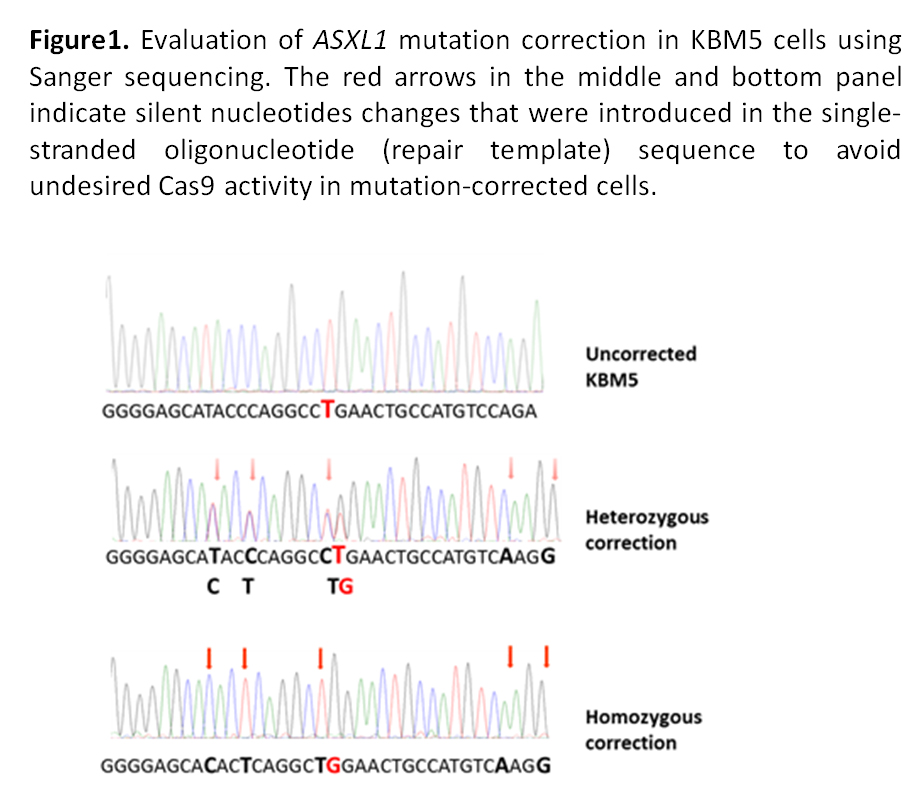
We used three synthetic guide RNAs (sgRNAs) targeting the genomic region overlapping the ASXL1 mutation and a single-stranded oligonucleotide as a repair template. We sequenced 1,027 colonies expanded from single FACS-sorted KBM5 cells to determine successful HDR-mediated correction of ASXL1 mutation. Each sgRNA generated heterozygous precise correction (yield 0.46-2%). Importantly, homozygous precise correction was observed for two sgRNAs (yield 1.13-1.63%). The frequency of correction obtained is consistent with previous studies using the CRISPR/Cas9 system for precise genome editing through HDR.
Results
Western blot analysis showed restoration of full-length ASXL1 protein expression in heterozygous and homozygous corrected KBM5 clones. ASXL1 plays an important role in Polycomb Repressive Complex 2 (PRC2) recruitment to specific target loci, including the HOXA cluster. Loss of ASXL1 is associated with increased expression of HOXA genes. We observed significant downregulation of HOXA5, 6, 7, 9, 10 and 13 in ASXL1 mutation-corrected KBM5 clones.
The PRC2 plays a critical role in the deposition of H3K27me3 histone repressive marks, and ASXL1 mutations are associated with H3K27me3 loss in myeloid cells. Western blots on purified histones showed a marked increase in global H3K27me3 levels in ASXL1 mutation-corrected KBM5 clones. ASXL1 forms a protein complex in vitro with the chromatin deubiquitinase BAP1. This interaction is reduced in cell lines carrying ASXL1 mutations. Immunoprecipitation studies showed that the interaction between ASXL1 and BAP1 was restored in ASXL1 mutation-corrected KBM5 clones.
Moreover, we observed growth suppression and a significant increase in the expression of the myeloid marker CD15 in ASXL1 mutation-corrected cells.
No sequence alterations were detected in any of the off-target sites examined in all ASXL1 mutation-corrected clones identified, suggesting high specificity of CRISPR/Cas9 system in our experiments.
Preliminary in vivo data show that mice xenografted with ASXL1 mutation-corrected KBM5 clones have a longer survival compared to mice xenografted with uncorrected KBM5 cells (p=0.046). A larger cohort is in progress.
Summary
In summary, we have corrected a specific ASXL1 point mutation in a leukemia cell line using CRISPR/Cas9 technology, resulting in restored protein function. This study presents a new strategy to illuminate the impact of oncogenic mutations on cellular function and may lay the foundations for a new approach to leukemia therapeutics (e.g. modified autoSCT). We provide proof-of-concept for gene correction via CRISPR/Cas9 technology in leukemic cells from patients with a myeloid malignancy.
Keyword(s): Chronic myeloid leukemia, Genetic, MDS, Mutation
Session topic: Novel actors in chronic myeloid leukemia biology
Type: Oral Presentation + travel grant
Presentation during EHA20: From 13.06.2015 12:00 to 13.06.2015 12:15
Location: Room Stolz 1
Background
The CRISPR/Cas9 genome editing system has recently emerged as a powerful tool for genome engineering, and has been used for gene correction in cultured cells from patients with monogenic hereditary defects. We reasoned that this technology could be employed to correct acquired gene mutations in human cancer cells.
Recurrent somatic mutations of the epigenetic modifier ASXL1 are common in myeloid malignancies, including myelodysplastic syndromes and chronic myeloid leukemia (CML). Loss-of-function ASXL1 mutations are strongly associated with a poor prognosis in these disorders. We studied the CML line KBM5 which lacks wild-type ASXL1 protein expression, due to a homozygous nonsense mutation (G710X) in the ASXL1 gene.
Aims
We used CRISPR/Cas9-mediated homology directed repair (HDR) to correct the ASXL1 mutation in KBM5 cells and performed functional studies to determine whether the wild-type function of ASXL1 was restored.
Methods
We used three synthetic guide RNAs (sgRNAs) targeting the genomic region overlapping the ASXL1 mutation and a single-stranded oligonucleotide as a repair template. We sequenced 1,027 colonies expanded from single FACS-sorted KBM5 cells to determine successful HDR-mediated correction of ASXL1 mutation. Each sgRNA generated heterozygous precise correction (yield 0.46-2%). Importantly, homozygous precise correction was observed for two sgRNAs (yield 1.13-1.63%). The frequency of correction obtained is consistent with previous studies using the CRISPR/Cas9 system for precise genome editing through HDR.
Results
Western blot analysis showed restoration of full-length ASXL1 protein expression in heterozygous and homozygous corrected KBM5 clones. ASXL1 plays an important role in Polycomb Repressive Complex 2 (PRC2) recruitment to specific target loci, including the HOXA cluster. Loss of ASXL1 is associated with increased expression of HOXA genes. We observed significant downregulation of HOXA5, 6, 7, 9, 10 and 13 in ASXL1 mutation-corrected KBM5 clones.
The PRC2 plays a critical role in the deposition of H3K27me3 histone repressive marks, and ASXL1 mutations are associated with H3K27me3 loss in myeloid cells. Western blots on purified histones showed a marked increase in global H3K27me3 levels in ASXL1 mutation-corrected KBM5 clones. ASXL1 forms a protein complex in vitro with the chromatin deubiquitinase BAP1. This interaction is reduced in cell lines carrying ASXL1 mutations. Immunoprecipitation studies showed that the interaction between ASXL1 and BAP1 was restored in ASXL1 mutation-corrected KBM5 clones.
Moreover, we observed growth suppression and a significant increase in the expression of the myeloid marker CD15 in ASXL1 mutation-corrected cells.
No sequence alterations were detected in any of the off-target sites examined in all ASXL1 mutation-corrected clones identified, suggesting high specificity of CRISPR/Cas9 system in our experiments.
Preliminary in vivo data show that mice xenografted with ASXL1 mutation-corrected KBM5 clones have a longer survival compared to mice xenografted with uncorrected KBM5 cells (p=0.046). A larger cohort is in progress.
Summary
In summary, we have corrected a specific ASXL1 point mutation in a leukemia cell line using CRISPR/Cas9 technology, resulting in restored protein function. This study presents a new strategy to illuminate the impact of oncogenic mutations on cellular function and may lay the foundations for a new approach to leukemia therapeutics (e.g. modified autoSCT). We provide proof-of-concept for gene correction via CRISPR/Cas9 technology in leukemic cells from patients with a myeloid malignancy.
Keyword(s): Chronic myeloid leukemia, Genetic, MDS, Mutation

Session topic: Novel actors in chronic myeloid leukemia biology


