Genomic Oncology

Contributions
Type: Oral Presentation
Presentation during EHA20: From 14.06.2015 08:45 to 14.06.2015 09:00
Location: Room Lehar 1 + 2
Background
Invasive Aspergillosis (IA) is a life-threatening infection caused by Aspergillus that mainly affects acute myelogenous leukemia and allogeneic stem cell transplantation (allo-SCT) patients. Recent studies suggest that immunomodulating single nucleotide polymorphisms (SNPs) may influence on the risk of developing the infection.
Aims
The purpose of this study was to assess whether 36 SNPs within 14 immune-related genes (IL4, IL4R, IL8, IL8RA, IL8RB, IL10, IL12A, IL12B, IL13, IFNg, IFNgR2, CCR5, MIF and VEGF) are associated with the risk of IA and whether a predictive model built with these variants might help to predict the disease risk.
Methods
We conducted a three-stage case-control association study of 742 high-risk patients, 146 of whom were diagnosed with proven or probable IA. This is one of the largest populations recruited so far to conduct genetic association studies.
Results
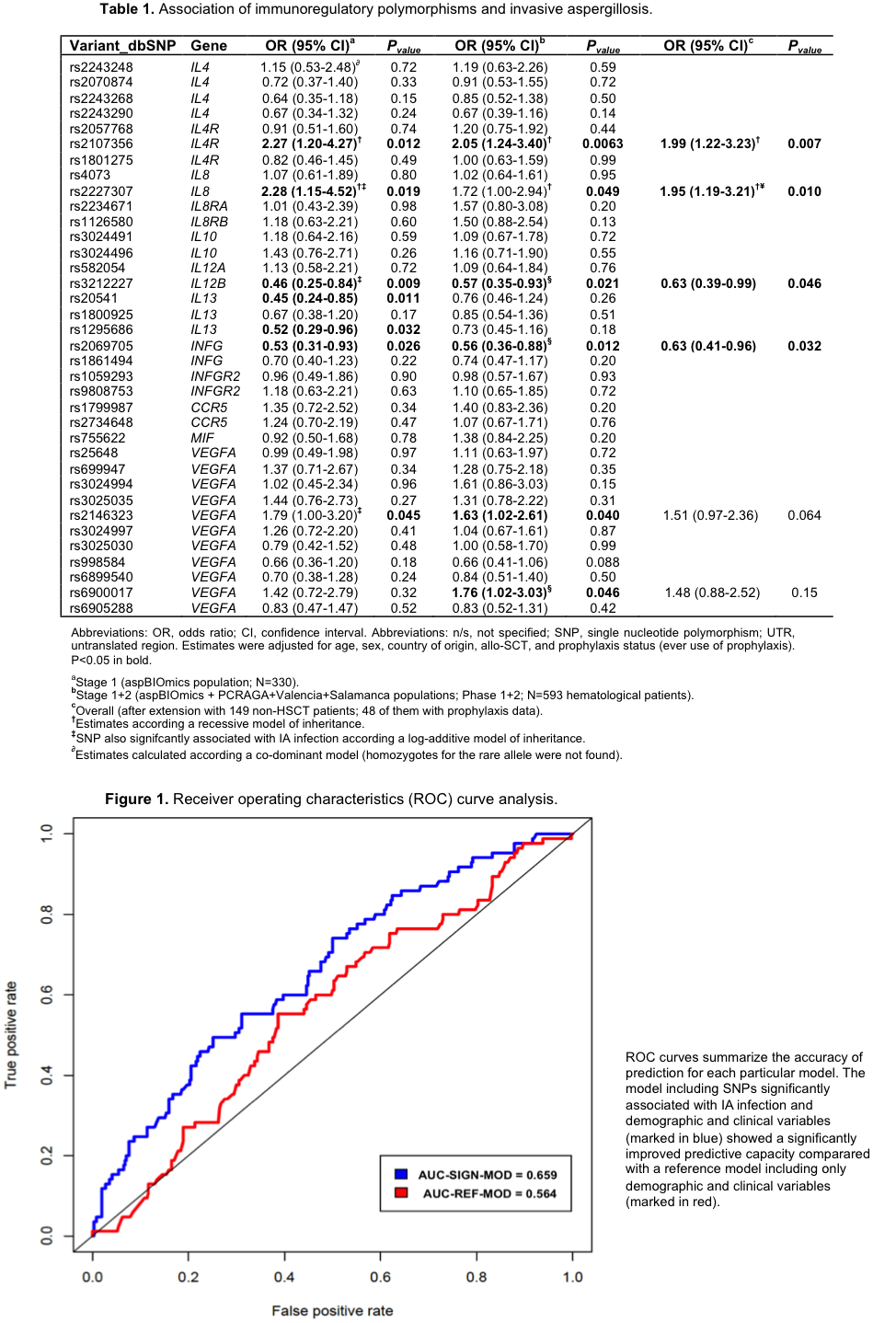
Overall, we found that the IL4Rrs2107356 and IL8rs2227307 SNPs were associated with an increased risk of IA (P=0.007 and P=0.010, respectively) whereas the IL12Brs3212227 and IFNgrs2069705 variants were significantly associated with a decreased risk of developing the disease (P=0.046 and P=0.032; Table 1). Importantly, an allogeneic stem cell transplantation (allo-SCT)-stratified analysis revealed that the effect observed for the IL4Rrs2107356 and IFNgrs2069705 SNPs was stronger in allo-SCT patients (P=0.0007 andP=0.0010, respectively) compared with those patients without transplantation. Although none of these associations remained significant after correction for multiple testing (P=0.0004), the association ofIL4Rrs2107356 and IFNgrs2069705 SNPs in allo-SCT patients remained marginally associated with the risk of IA infection. In vitro stimulation assays confirmed a relevant role of the IL4R2107356 and IL12Brs3212227SNPs in regulating IL4R and IL12 levels. We found that CD19+B lymphocytes from carriers of theIL4R2107356A/A mutant genotype (n=7) tended to have an increased expression of IL4R protein when compared with those B-cells from subjects harbouring the wild-type allele (n=7; P=0.08). Although carriers of the IL4R2107356A/A genotype also tended to have higher levels of IL4R in T-cells and monocytes, a substantial correlation could only be detected in B-lymphocytes, a cell subset where IL4R is highly expressed. We also confirmed that carriers of the IL12Brs3212227C allele showed an increased production of IL12p70 after 24 and 48h of incubation with zymosan alone or in combination with LPS when compared with carriers of the IL12Brs3212227A/A genotype (IL12BMUT-ZYM-24h=44.0±8.5 vs. IL12BWT-ZYM-24h=30.1±2.2, P=0.087 and IL12BMUT-ZYM-48h=81.9±9.4 vs. IL12BWT-ZYM-48h=26.8±5.8, P=0.006 and IL12BWT-ZYM+LPS-24h=34.6±3.0 vs. IL12BMUT-ZYM+LPS-24h=73.3±4.0,P=0.0017). In addition, we found that patients harbouring the IL12Brs3212227C allele showed a substantially increased level of IFNg after 48h of incubation with Zymosan when compared with those subjects harbouring the wild type genotype (IFNgMUT-ZYM-48h=501.4±38.1 vs. IFNgWT-ZYM-48h=183.5±83.0, P=0.06). Finally, a predictive analysis also confirmed that a prediction model including SNPs significantly associated with IA showed a substantial improvement in the discriminatory ability to predict the disease when compared with a reference model including only demographic and clinical variables (AUC=0.659 vs. AUC=0.564; Figure 1).
Summary
These findings suggest that SNPs within immuno-modulating genes influence on the risk of developing IA infection and might be used to predict the disease risk and to implement risk-adapted prophylaxis strategies.
Keyword(s): IL-12, Invasive aspergillosis, Polymorphism, Prediction
Session topic: Stem cell transplantation: Clinical 3
Type: Oral Presentation
Presentation during EHA20: From 14.06.2015 08:45 to 14.06.2015 09:00
Location: Room Lehar 1 + 2
Background
Invasive Aspergillosis (IA) is a life-threatening infection caused by Aspergillus that mainly affects acute myelogenous leukemia and allogeneic stem cell transplantation (allo-SCT) patients. Recent studies suggest that immunomodulating single nucleotide polymorphisms (SNPs) may influence on the risk of developing the infection.
Aims
The purpose of this study was to assess whether 36 SNPs within 14 immune-related genes (IL4, IL4R, IL8, IL8RA, IL8RB, IL10, IL12A, IL12B, IL13, IFNg, IFNgR2, CCR5, MIF and VEGF) are associated with the risk of IA and whether a predictive model built with these variants might help to predict the disease risk.
Methods
We conducted a three-stage case-control association study of 742 high-risk patients, 146 of whom were diagnosed with proven or probable IA. This is one of the largest populations recruited so far to conduct genetic association studies.
Results
Overall, we found that the IL4Rrs2107356 and IL8rs2227307 SNPs were associated with an increased risk of IA (P=0.007 and P=0.010, respectively) whereas the IL12Brs3212227 and IFNgrs2069705 variants were significantly associated with a decreased risk of developing the disease (P=0.046 and P=0.032; Table 1). Importantly, an allogeneic stem cell transplantation (allo-SCT)-stratified analysis revealed that the effect observed for the IL4Rrs2107356 and IFNgrs2069705 SNPs was stronger in allo-SCT patients (P=0.0007 andP=0.0010, respectively) compared with those patients without transplantation. Although none of these associations remained significant after correction for multiple testing (P=0.0004), the association ofIL4Rrs2107356 and IFNgrs2069705 SNPs in allo-SCT patients remained marginally associated with the risk of IA infection. In vitro stimulation assays confirmed a relevant role of the IL4R2107356 and IL12Brs3212227SNPs in regulating IL4R and IL12 levels. We found that CD19+B lymphocytes from carriers of theIL4R2107356A/A mutant genotype (n=7) tended to have an increased expression of IL4R protein when compared with those B-cells from subjects harbouring the wild-type allele (n=7; P=0.08). Although carriers of the IL4R2107356A/A genotype also tended to have higher levels of IL4R in T-cells and monocytes, a substantial correlation could only be detected in B-lymphocytes, a cell subset where IL4R is highly expressed. We also confirmed that carriers of the IL12Brs3212227C allele showed an increased production of IL12p70 after 24 and 48h of incubation with zymosan alone or in combination with LPS when compared with carriers of the IL12Brs3212227A/A genotype (IL12BMUT-ZYM-24h=44.0±8.5 vs. IL12BWT-ZYM-24h=30.1±2.2, P=0.087 and IL12BMUT-ZYM-48h=81.9±9.4 vs. IL12BWT-ZYM-48h=26.8±5.8, P=0.006 and IL12BWT-ZYM+LPS-24h=34.6±3.0 vs. IL12BMUT-ZYM+LPS-24h=73.3±4.0,P=0.0017). In addition, we found that patients harbouring the IL12Brs3212227C allele showed a substantially increased level of IFNg after 48h of incubation with Zymosan when compared with those subjects harbouring the wild type genotype (IFNgMUT-ZYM-48h=501.4±38.1 vs. IFNgWT-ZYM-48h=183.5±83.0, P=0.06). Finally, a predictive analysis also confirmed that a prediction model including SNPs significantly associated with IA showed a substantial improvement in the discriminatory ability to predict the disease when compared with a reference model including only demographic and clinical variables (AUC=0.659 vs. AUC=0.564; Figure 1).
Summary
These findings suggest that SNPs within immuno-modulating genes influence on the risk of developing IA infection and might be used to predict the disease risk and to implement risk-adapted prophylaxis strategies.
Keyword(s): IL-12, Invasive aspergillosis, Polymorphism, Prediction

Session topic: Stem cell transplantation: Clinical 3


