
Contributions
Type: Oral Presentation
Presentation during EHA20: From 13.06.2015 16:15 to 13.06.2015 16:30
Location: Room Stolz 1
Background
Inherited platelet disorders (IPDs) cause bleeding diathesis with varying severity. Due to their rarity and extreme heterogeneity, the diagnosis is complex, poorly standardized, time consuming and expensive. Based on clinical and laboratory findings, gene mutations may be assessed by means of conventional Sanger sequencing to diagnose the disease. However, in many cases a proper diagnosis and the genetic cause for the disease remain unknown. Next generation sequencing (NGS) enables the simultaneous analysis of large groups of candidate genes in IPDs and may be useful for rapid genetic diagnosis and allow optimal disease classification and understanding of the pathogenesis of the disease.
Aims
To improve the diagnosis of IPDs by using a next generation sequencing panel.
Methods
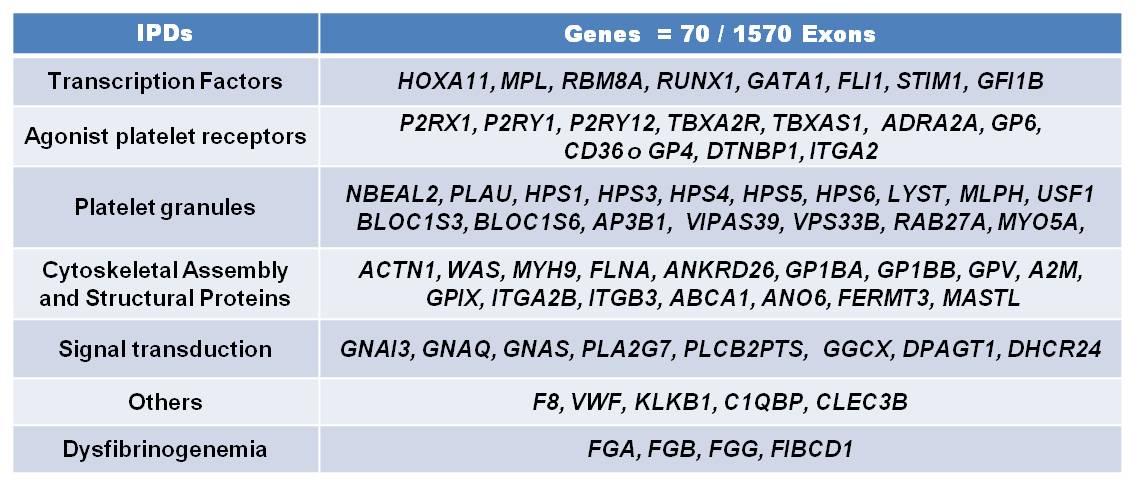
Forty patients with suspected IPDs were analysed by NGS panel to identify a pathogenic mutation causes them. Patients had abnormal bleeding symptoms, normal coagulation factors levels, platelet function testing pathological or thrombocytopenia without evidence of acquired causes. Two groups were established, first group, 19 patients with specific phenotype and 21 without a proper diagnosis or unknown phenotype. For the 70 genes associated with IPDs (figure1) baits were generated to tile 400 kb DNA, corresponding to the exons and splice sites of all known transcripts of the candidate genes. The bait library was tested by enriching candidate genes from 50 ng DNA using the Nextera Rapid Custom Enrichment system followed by massive parallel sequencing (Illumina).
Results
Genotype were determined in 16 of 19 patients with specific phenotype, therefore IPDs diagnosis were confirmed: 4 Von Willebrand Disease (VWD), 1 Glanzmann Thrombastenia (GT), 4 Bernard Soulier syndrome (BSS), 2 MYH9-related disease, 1 Hermansky Pudlack Syndrome, 1 Prekalicrein defiency, 2 Dysfibrinogenemia, 1 Signalling defect (GP6). Patients with unknown phenotype, were subdivided in patients with or without thrombocytopenia. In patients with thrombocytopenia diagnosis were indentified in 7/10: 2 BSS (GP1BA; c.463C>; c.1280_1291delCCTCAGAGCCCG); VWD-PT (GP1BA; c.733G>A); Bening Mediterranean macrothrombocytopenia (GP1BB; c.1A>T), Gray Platelet syndrome (NBEAL2; c.3422_3423insC), Wiskott-Aldrich Syndrome (WAS; c.802delC) and Thrombocytopenia FLNA-related (FLNA; c.3695C>T). In IPD patients without thrombocytopenia diagnosis were identified in 6/11: GT (ITGA2B; c.2063C>T); VWD 2N (VWF; c.2561G>A), mild Hemophilia A (F8, c.6623A>G), GATA-related disease (GATA1; C.865C>T), RUNX1-related disease (RUNX1; c.167T>T), and Defect ADP receptor (P2YR12, c.835G>A). No mutations were found in 3 patients with “Aspirin-like” syndrome. In fact, genotype were determined in 13 of 21; and 22 mutations have not previously been described in the literature. Types of mutations described were 33 missense and 8 frameshift (3 insertion, 5 deletion). All mutations identified by NGS were confirmed by Sanger sequencing. Once, diagnosis was determined, functional testing, flow cytometry and electron microscopy were done to establish a proper diagnosis of IPDs.
Summary
NGS by amplicon-based capture enables a fast diagnosis of IPDs. This tool cold be used ina clinical setting to facilitate the diagnosis of IPD displaying an unclear phenoptype.
Keyword(s): Diagnosis, Inherited platelet disorders, Mutation analysis
Session topic: Platelet and bleeding disorders
Type: Oral Presentation
Presentation during EHA20: From 13.06.2015 16:15 to 13.06.2015 16:30
Location: Room Stolz 1
Background
Inherited platelet disorders (IPDs) cause bleeding diathesis with varying severity. Due to their rarity and extreme heterogeneity, the diagnosis is complex, poorly standardized, time consuming and expensive. Based on clinical and laboratory findings, gene mutations may be assessed by means of conventional Sanger sequencing to diagnose the disease. However, in many cases a proper diagnosis and the genetic cause for the disease remain unknown. Next generation sequencing (NGS) enables the simultaneous analysis of large groups of candidate genes in IPDs and may be useful for rapid genetic diagnosis and allow optimal disease classification and understanding of the pathogenesis of the disease.
Aims
To improve the diagnosis of IPDs by using a next generation sequencing panel.
Methods
Forty patients with suspected IPDs were analysed by NGS panel to identify a pathogenic mutation causes them. Patients had abnormal bleeding symptoms, normal coagulation factors levels, platelet function testing pathological or thrombocytopenia without evidence of acquired causes. Two groups were established, first group, 19 patients with specific phenotype and 21 without a proper diagnosis or unknown phenotype. For the 70 genes associated with IPDs (figure1) baits were generated to tile 400 kb DNA, corresponding to the exons and splice sites of all known transcripts of the candidate genes. The bait library was tested by enriching candidate genes from 50 ng DNA using the Nextera Rapid Custom Enrichment system followed by massive parallel sequencing (Illumina).
Results
Genotype were determined in 16 of 19 patients with specific phenotype, therefore IPDs diagnosis were confirmed: 4 Von Willebrand Disease (VWD), 1 Glanzmann Thrombastenia (GT), 4 Bernard Soulier syndrome (BSS), 2 MYH9-related disease, 1 Hermansky Pudlack Syndrome, 1 Prekalicrein defiency, 2 Dysfibrinogenemia, 1 Signalling defect (GP6). Patients with unknown phenotype, were subdivided in patients with or without thrombocytopenia. In patients with thrombocytopenia diagnosis were indentified in 7/10: 2 BSS (GP1BA; c.463C>; c.1280_1291delCCTCAGAGCCCG); VWD-PT (GP1BA; c.733G>A); Bening Mediterranean macrothrombocytopenia (GP1BB; c.1A>T), Gray Platelet syndrome (NBEAL2; c.3422_3423insC), Wiskott-Aldrich Syndrome (WAS; c.802delC) and Thrombocytopenia FLNA-related (FLNA; c.3695C>T). In IPD patients without thrombocytopenia diagnosis were identified in 6/11: GT (ITGA2B; c.2063C>T); VWD 2N (VWF; c.2561G>A), mild Hemophilia A (F8, c.6623A>G), GATA-related disease (GATA1; C.865C>T), RUNX1-related disease (RUNX1; c.167T>T), and Defect ADP receptor (P2YR12, c.835G>A). No mutations were found in 3 patients with “Aspirin-like” syndrome. In fact, genotype were determined in 13 of 21; and 22 mutations have not previously been described in the literature. Types of mutations described were 33 missense and 8 frameshift (3 insertion, 5 deletion). All mutations identified by NGS were confirmed by Sanger sequencing. Once, diagnosis was determined, functional testing, flow cytometry and electron microscopy were done to establish a proper diagnosis of IPDs.
Summary
NGS by amplicon-based capture enables a fast diagnosis of IPDs. This tool cold be used ina clinical setting to facilitate the diagnosis of IPD displaying an unclear phenoptype.
Keyword(s): Diagnosis, Inherited platelet disorders, Mutation analysis

Session topic: Platelet and bleeding disorders


