LEUKAEMIA-ASSOCIATED SOMATIC MUTATIONS DRIVE DISTINCT PATTERNS OF AGE-RELATED CLONAL HAEMOPOIESIS
(Abstract release date: 05/21/15)
EHA Library. McKerrell T. 06/13/15; 103060; S452
Disclosure(s): Wellcome Trust Sanger InstituteHaematological Cancer Genetics

Thomas McKerrell
Contributions
Contributions
Abstract
Abstract: S452
Type: Oral Presentation + travel grant
Presentation during EHA20: From 13.06.2015 11:45 to 13.06.2015 12:00
Location: Room Lehar 1 + 2
Background
Clonal haemopoiesis driven by leukaemia-associated gene mutations can occur without evidence of a blood disorder.
Aims
To investigate the incidence, target genes and age distribution of age-related clonal haemopoiesis (ARCH).
Methods
We performed targeted ultra-deep re-sequencing for hotspot mutations at 15 gene loci recurrently mutated in myeloid malignancies in the blood DNA of 4219 individuals including a large number of elderly people.To do this we developed and validated a robust methodology, employing barcoded multiplex PCR of mutational hotspots followed by next-generation sequencing (MiSeq) and bioinformatic analysis. This reliably detected mutation-associated circulating blood cell clones with a variant allele fraction (VAF) ≥ 0.008 (0.8%).
Results
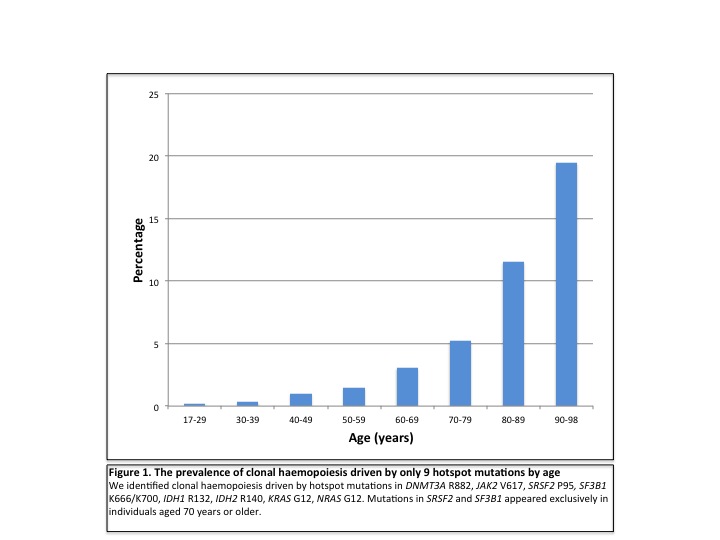
Using only the hotspots studied, we identified clonal haemopoiesis in 0.8% of individuals under 60, rising to 19.5% of those ≥90 years; predicting that clonal haemopoiesis is much more prevalent overall than was previously realized. Indeed, using our findings as a basis for projection, we estimate the overall prevalence of age related clonal haemopoiesis (ARCH) to be >70% in those older than 90 years. DNMT3A-R882 mutations were most common and, although their incidence increased with age, were found in individuals as young as 25 years. By contrast mutations affecting spliceosome genes SF3B1 and SRSF2, closely associated with the myelodysplastic syndromes, were only identified in those aged >70 years, with several individuals harboring more than one such mutation.This indicates that spliceosome gene mutations drive clonal expansion under selection pressures operating in the ageing haemopoietic system and explains the high incidence of clonal disorders associated with these mutations in advanced old age. Finally, despite using a very sensitive method and a mutation-calling script written specifically for this purpose, no samples with NPM1 mutations of VAF ≥ 0.008 were identified.
Summary
Our results demonstrate that the incidence of clonal haemopoiesis is much higher than suggested by exome sequencing studies, that splicesome gene mutations drive clonal outgrowth primarily in the context of an aging haematopoietic compartment, and that NPM1 mutations do not drive ARCH, indicating that their acquisition is closely associated with frank leukemia
Keyword(s): Age, Clonality, Leukemogenesis, Somatic mutation
Session topic: Molecular markers in AML
Type: Oral Presentation + travel grant
Presentation during EHA20: From 13.06.2015 11:45 to 13.06.2015 12:00
Location: Room Lehar 1 + 2
Background
Clonal haemopoiesis driven by leukaemia-associated gene mutations can occur without evidence of a blood disorder.
Aims
To investigate the incidence, target genes and age distribution of age-related clonal haemopoiesis (ARCH).
Methods
We performed targeted ultra-deep re-sequencing for hotspot mutations at 15 gene loci recurrently mutated in myeloid malignancies in the blood DNA of 4219 individuals including a large number of elderly people.To do this we developed and validated a robust methodology, employing barcoded multiplex PCR of mutational hotspots followed by next-generation sequencing (MiSeq) and bioinformatic analysis. This reliably detected mutation-associated circulating blood cell clones with a variant allele fraction (VAF) ≥ 0.008 (0.8%).
Results
Using only the hotspots studied, we identified clonal haemopoiesis in 0.8% of individuals under 60, rising to 19.5% of those ≥90 years; predicting that clonal haemopoiesis is much more prevalent overall than was previously realized. Indeed, using our findings as a basis for projection, we estimate the overall prevalence of age related clonal haemopoiesis (ARCH) to be >70% in those older than 90 years. DNMT3A-R882 mutations were most common and, although their incidence increased with age, were found in individuals as young as 25 years. By contrast mutations affecting spliceosome genes SF3B1 and SRSF2, closely associated with the myelodysplastic syndromes, were only identified in those aged >70 years, with several individuals harboring more than one such mutation.This indicates that spliceosome gene mutations drive clonal expansion under selection pressures operating in the ageing haemopoietic system and explains the high incidence of clonal disorders associated with these mutations in advanced old age. Finally, despite using a very sensitive method and a mutation-calling script written specifically for this purpose, no samples with NPM1 mutations of VAF ≥ 0.008 were identified.
Summary
Our results demonstrate that the incidence of clonal haemopoiesis is much higher than suggested by exome sequencing studies, that splicesome gene mutations drive clonal outgrowth primarily in the context of an aging haematopoietic compartment, and that NPM1 mutations do not drive ARCH, indicating that their acquisition is closely associated with frank leukemia
Keyword(s): Age, Clonality, Leukemogenesis, Somatic mutation

Session topic: Molecular markers in AML
Abstract: S452
Type: Oral Presentation + travel grant
Presentation during EHA20: From 13.06.2015 11:45 to 13.06.2015 12:00
Location: Room Lehar 1 + 2
Background
Clonal haemopoiesis driven by leukaemia-associated gene mutations can occur without evidence of a blood disorder.
Aims
To investigate the incidence, target genes and age distribution of age-related clonal haemopoiesis (ARCH).
Methods
We performed targeted ultra-deep re-sequencing for hotspot mutations at 15 gene loci recurrently mutated in myeloid malignancies in the blood DNA of 4219 individuals including a large number of elderly people.To do this we developed and validated a robust methodology, employing barcoded multiplex PCR of mutational hotspots followed by next-generation sequencing (MiSeq) and bioinformatic analysis. This reliably detected mutation-associated circulating blood cell clones with a variant allele fraction (VAF) ≥ 0.008 (0.8%).
Results
Using only the hotspots studied, we identified clonal haemopoiesis in 0.8% of individuals under 60, rising to 19.5% of those ≥90 years; predicting that clonal haemopoiesis is much more prevalent overall than was previously realized. Indeed, using our findings as a basis for projection, we estimate the overall prevalence of age related clonal haemopoiesis (ARCH) to be >70% in those older than 90 years. DNMT3A-R882 mutations were most common and, although their incidence increased with age, were found in individuals as young as 25 years. By contrast mutations affecting spliceosome genes SF3B1 and SRSF2, closely associated with the myelodysplastic syndromes, were only identified in those aged >70 years, with several individuals harboring more than one such mutation.This indicates that spliceosome gene mutations drive clonal expansion under selection pressures operating in the ageing haemopoietic system and explains the high incidence of clonal disorders associated with these mutations in advanced old age. Finally, despite using a very sensitive method and a mutation-calling script written specifically for this purpose, no samples with NPM1 mutations of VAF ≥ 0.008 were identified.
Summary
Our results demonstrate that the incidence of clonal haemopoiesis is much higher than suggested by exome sequencing studies, that splicesome gene mutations drive clonal outgrowth primarily in the context of an aging haematopoietic compartment, and that NPM1 mutations do not drive ARCH, indicating that their acquisition is closely associated with frank leukemia
Keyword(s): Age, Clonality, Leukemogenesis, Somatic mutation

Session topic: Molecular markers in AML
Type: Oral Presentation + travel grant
Presentation during EHA20: From 13.06.2015 11:45 to 13.06.2015 12:00
Location: Room Lehar 1 + 2
Background
Clonal haemopoiesis driven by leukaemia-associated gene mutations can occur without evidence of a blood disorder.
Aims
To investigate the incidence, target genes and age distribution of age-related clonal haemopoiesis (ARCH).
Methods
We performed targeted ultra-deep re-sequencing for hotspot mutations at 15 gene loci recurrently mutated in myeloid malignancies in the blood DNA of 4219 individuals including a large number of elderly people.To do this we developed and validated a robust methodology, employing barcoded multiplex PCR of mutational hotspots followed by next-generation sequencing (MiSeq) and bioinformatic analysis. This reliably detected mutation-associated circulating blood cell clones with a variant allele fraction (VAF) ≥ 0.008 (0.8%).
Results
Using only the hotspots studied, we identified clonal haemopoiesis in 0.8% of individuals under 60, rising to 19.5% of those ≥90 years; predicting that clonal haemopoiesis is much more prevalent overall than was previously realized. Indeed, using our findings as a basis for projection, we estimate the overall prevalence of age related clonal haemopoiesis (ARCH) to be >70% in those older than 90 years. DNMT3A-R882 mutations were most common and, although their incidence increased with age, were found in individuals as young as 25 years. By contrast mutations affecting spliceosome genes SF3B1 and SRSF2, closely associated with the myelodysplastic syndromes, were only identified in those aged >70 years, with several individuals harboring more than one such mutation.This indicates that spliceosome gene mutations drive clonal expansion under selection pressures operating in the ageing haemopoietic system and explains the high incidence of clonal disorders associated with these mutations in advanced old age. Finally, despite using a very sensitive method and a mutation-calling script written specifically for this purpose, no samples with NPM1 mutations of VAF ≥ 0.008 were identified.
Summary
Our results demonstrate that the incidence of clonal haemopoiesis is much higher than suggested by exome sequencing studies, that splicesome gene mutations drive clonal outgrowth primarily in the context of an aging haematopoietic compartment, and that NPM1 mutations do not drive ARCH, indicating that their acquisition is closely associated with frank leukemia
Keyword(s): Age, Clonality, Leukemogenesis, Somatic mutation

Session topic: Molecular markers in AML
{{ help_message }}
{{filter}}


