LOSANTE HOSPITAL FOR CHILDREN WITH LEUKEMIA

Contributions
Type: Publication Only
Background
CD22, a B cell specific transmembrane protein, have commonly seen in hematological malignancies and plays a role in B cell survival, proliferation, and differentiation via B-cell receptor (BCR). BCR is an inhibitory co-receptor of B cells and B cell precursors that acts as a negative regulator of multiple signal transduction pathways critical for B cells. CD22 signaling is mediated through interactions with kinases and phosphatases which connect the cytoplasmic domain via phosphorylated tyrosine residues that locate within TAM and TIM motifs.
CD22 protein SHP-1 (Src homology 2 domain-containing tyrosine phosphatase) function deficient mice showed disruption of the SHP-1 signaling network that can result in defective maturation in development of B-cell lymphoproliferative state and apoptosis. Studies of human CD22 demonstrated that its variations could cause CD22 molecule with inadequate functional and disturbance of mRNA expression.
CD22 is expressed in 90% of chronic lymphoblastic leukemia patients, in 60-70% of B cell lymphoma patients, in 100% of hairy cell leukemia patients, in 96% of childhood acute lymphoblastic leukemia patients. Precursor B Cell Acute Lymphoblastic Leukemia (precursor B-ALL) the largest subset of acute lymphoblastic leukemia (ALL), is the most common form of childhood cancer.
B cell expression of CD22 makes this a great target for the treatment of leukemia. CD22 that is located on chromosome 19q24 contains 14 exons. Studies by using DNA sequencing method were detected several homozygous mutations (transversions/transitions, deletions, and insertions) between exon 12 and intron 13 in CD22 gene and this region was called “mutational hotspot”.
Aims
In this study we aimed to screen exon 9, 10 and 12 of CD22 gene that is mutational hotspot region in children with precursor B-ALL and to find possible genetic variations for molecular hematological analysis in childhood ALL.
Methods
In this study we aimed to screen exon 9, 10 and 12 of CD22 gene that is mutational hotspot region in children with precursor B-ALL and to find possible genetic variations for molecular hematological analysis in childhood ALL.
Study population consisted of 115 patients aged between 1 and 15 years who were admitted to Lösante Hospital with the diagnosis of precursor B-ALL. Blood samples were collected with EDTA-containing tubes and DNA was extracted from peripheral blood and bone marrow leukocytes with MagNA Pure automatic DNA isolation instrument (Roche Diagnostics, Manheim, Germany). Amplification of gene was performed by PCR and all samples were screened for the variants by SSCP. Samples showing band shifts were directly sequenced on an automated sequencer (Beckman Coulter, USA).
Results
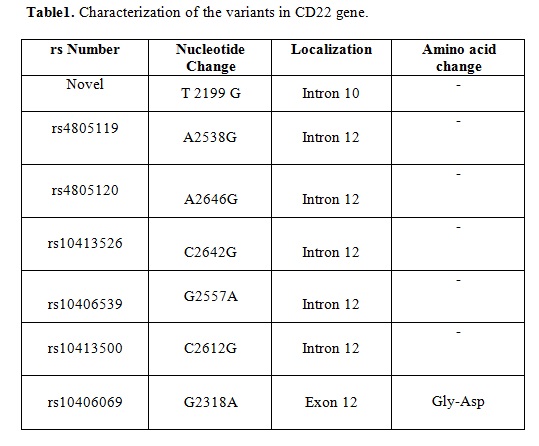
We detected 7 variants in CD22 gene in our study population. There were no variants at exon 9. We report a novel mutation at exon 10 of CD22 (T2199G). The most of CD22 variants were described in exon 12; intron A2538G (rs4805119), intron A2646G (rs4805120), intron C2642G (rs10413526); intron G2557A (rs10406539); intron C2612G (rs10413500); exon G2318A (rs10406069). Table 1 shows summary of CD22 variant profiles.
Summary
The detected variants in this study seem to be the first screening results of studied gene in childhood precursor B-ALL patients in our country. Identified variants in the CD22 exons encoding the cytoplasmic domain could disturb mRNA stability and splicing mechanism of gene. Thus, a truncated CD22 protein or reduced expression levels of an intact protein might lead to development of leukemia. These results need to be confirmed by further exons on a larger number of patients.
Keyword(s): ALL, Children, Mutation analysis
Session topic: Publication Only
Type: Publication Only
Background
CD22, a B cell specific transmembrane protein, have commonly seen in hematological malignancies and plays a role in B cell survival, proliferation, and differentiation via B-cell receptor (BCR). BCR is an inhibitory co-receptor of B cells and B cell precursors that acts as a negative regulator of multiple signal transduction pathways critical for B cells. CD22 signaling is mediated through interactions with kinases and phosphatases which connect the cytoplasmic domain via phosphorylated tyrosine residues that locate within TAM and TIM motifs.
CD22 protein SHP-1 (Src homology 2 domain-containing tyrosine phosphatase) function deficient mice showed disruption of the SHP-1 signaling network that can result in defective maturation in development of B-cell lymphoproliferative state and apoptosis. Studies of human CD22 demonstrated that its variations could cause CD22 molecule with inadequate functional and disturbance of mRNA expression.
CD22 is expressed in 90% of chronic lymphoblastic leukemia patients, in 60-70% of B cell lymphoma patients, in 100% of hairy cell leukemia patients, in 96% of childhood acute lymphoblastic leukemia patients. Precursor B Cell Acute Lymphoblastic Leukemia (precursor B-ALL) the largest subset of acute lymphoblastic leukemia (ALL), is the most common form of childhood cancer.
B cell expression of CD22 makes this a great target for the treatment of leukemia. CD22 that is located on chromosome 19q24 contains 14 exons. Studies by using DNA sequencing method were detected several homozygous mutations (transversions/transitions, deletions, and insertions) between exon 12 and intron 13 in CD22 gene and this region was called “mutational hotspot”.
Aims
In this study we aimed to screen exon 9, 10 and 12 of CD22 gene that is mutational hotspot region in children with precursor B-ALL and to find possible genetic variations for molecular hematological analysis in childhood ALL.
Methods
In this study we aimed to screen exon 9, 10 and 12 of CD22 gene that is mutational hotspot region in children with precursor B-ALL and to find possible genetic variations for molecular hematological analysis in childhood ALL.
Study population consisted of 115 patients aged between 1 and 15 years who were admitted to Lösante Hospital with the diagnosis of precursor B-ALL. Blood samples were collected with EDTA-containing tubes and DNA was extracted from peripheral blood and bone marrow leukocytes with MagNA Pure automatic DNA isolation instrument (Roche Diagnostics, Manheim, Germany). Amplification of gene was performed by PCR and all samples were screened for the variants by SSCP. Samples showing band shifts were directly sequenced on an automated sequencer (Beckman Coulter, USA).
Results
We detected 7 variants in CD22 gene in our study population. There were no variants at exon 9. We report a novel mutation at exon 10 of CD22 (T2199G). The most of CD22 variants were described in exon 12; intron A2538G (rs4805119), intron A2646G (rs4805120), intron C2642G (rs10413526); intron G2557A (rs10406539); intron C2612G (rs10413500); exon G2318A (rs10406069). Table 1 shows summary of CD22 variant profiles.
Summary
The detected variants in this study seem to be the first screening results of studied gene in childhood precursor B-ALL patients in our country. Identified variants in the CD22 exons encoding the cytoplasmic domain could disturb mRNA stability and splicing mechanism of gene. Thus, a truncated CD22 protein or reduced expression levels of an intact protein might lead to development of leukemia. These results need to be confirmed by further exons on a larger number of patients.
Keyword(s): ALL, Children, Mutation analysis

Session topic: Publication Only


