
Contributions
Type: Publication Only
Background
JAK2 and MPL mutation play an important role in diagnosis of myeloid proliferative neoplasm (MPN). Another key marker in the molecular diagnosis of MPNs, mutations in CALR, was recently reported. Known mutations are all located at exon 9 and they are somatic mutations of deletion or insertions; the two commonest abnormalities account for 80-90% of CALR mutations, whereas lesions are highly heterogeneous in the remaining 10-20%.
Aims
Almost previous reports analyzed CALR mutations using Sanger sequencing. However, sequencing method can’t be used easily in every laboratory condition. Authors want to make an easy system to detect CALR mutations which is available in small laboratory without sequencing machine. We investigated the screening PCR to detect CALR mutations in Korean patients with thrombocytosis and compared the results with Sanger sequencing.
Methods
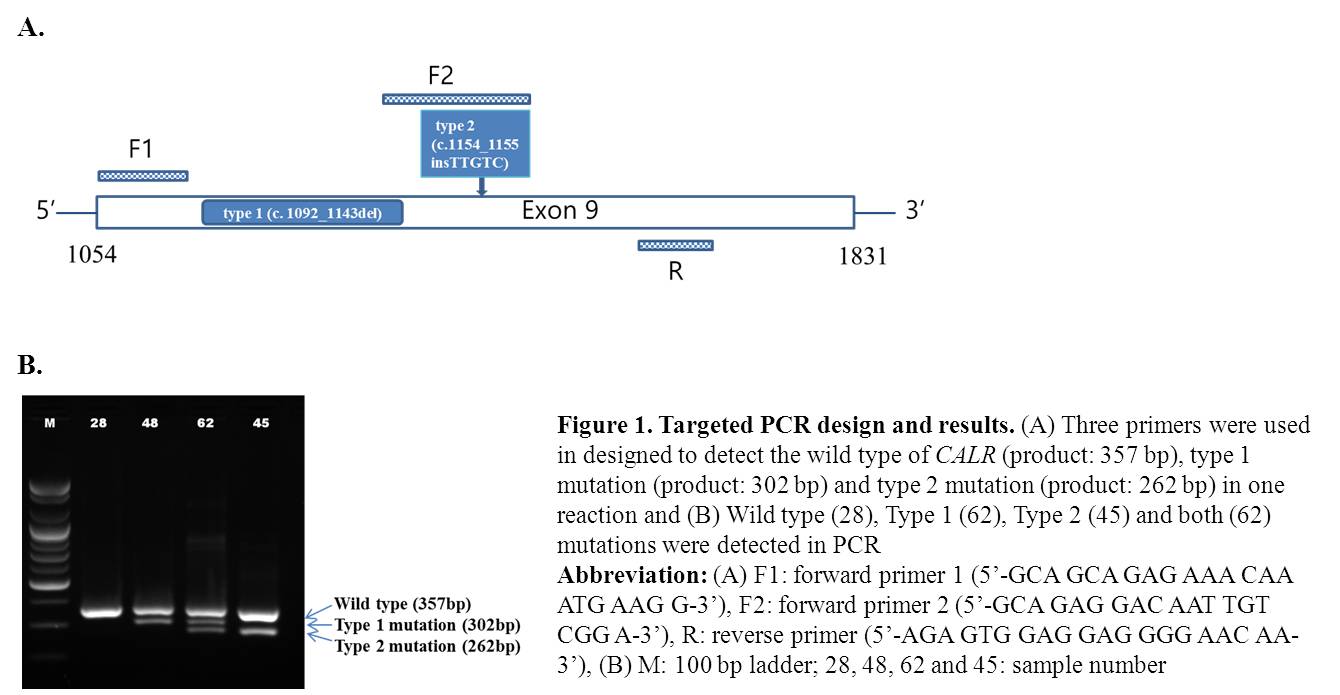
Bone marrow DNA samples were obtained from 83 patients with thrombocytosis. Screening PCR primer sets were designed to detect the wild type (product: 357 bp), type 1 mutation (product: 302 bp) and type 2 mutation (product: 262 bp) in one reaction (Figure 1). And Sanger sequencing was done in hot spot region.
Results
JAK2V617F was detected in 36 patients (33 ET, 1 PMF, 1 MPN, U, and 1 MDS/MPN, U) (Table 1). Two kinds of CALR mutation, type 1 (c.1092_1143del) and type 2 (c.1154_1155insTTGTC) were detected in 16 ET and 1 PMF patients. In ET group, mutational frequencies were 62.3% for JAK2V617F and, 30.2% for CALR, 5.7% for dual positive and 13.2% for dual negative. Among the ET patients without JAK2V617F, frequency of CALR mutation was 80%. The frequency of type 2 CALR mutation (8/16, 50.0%) was higher than type 1 (6/16, 37.5%). Interestingly, one patient with both mutations (1/16, 6.3%) was identified and novel mutation pattern (c.1123_1132delinsTGC) was detected by only Sanger sequencing. Any mutation was not detected in reactive disease group.
Table 1. The frequency of JAK2 and CALR mutations in different patient groups
Disease | ET | PMF | MDS/MPN, U | MPN, U | HES | Reactive | Total |
N=53 | N=3 | N=3 | N=2 | N=1 | N=21 | N=83 | |
JAK2 mutation, N (%) | 33 (62.3%) | 1 (33.3%) | 1 (33.3%) | 1 (50.0%) | 0 (0.0%) | 0 (0.0%) | 36 (43.4%) |
CALR mutation, N (%) | 16 (30.2%)* | 1 (33.3%) | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) | 17 (20.5%) |
Dual positive, N (%) | 3 (5.7%) | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) | 3 (3.6%) |
Dual negative, N (%) | 7 (13.2%) | 1 (33.3%) | 2 (66.7%) | 1 (50.0%) | 1 (100.0%) | 21 (100.0%) | 34 (41.0%) |
CALR mutation (%) | 80% | 50% |
*Type 1: 6 patients (37.5%); Type 2: 8 patients (50.0%); Type 1 & Type 2: 1 patient (6.3%) and novel mutation: 1 patient (6.3%)
Abbreviation: ET; essential thrombocythemia, PMF; primary myelofibrosis, MDS/MPN,U; myelodysplastic syndrome/myeloproliferative neoplasm, unclassifiable, MPN,U; myeloproliferative neoplasm, unclassifiable, HES; hypereosonophilic syndrome, Reactive; reactive disease group with non-hematologic malignancy
Summary
Screening PCR could detect the 94.1% (16/17) of mutation cases, and this method showed the concordant result with Sanger sequencing in the case of type 1 and type 2 mutations. One novel mutation was only identified with Sanger sequencing. Our system can’t detect single base pair substitution or small base insertions like the mutation of this case; however, the frequency of these mutations is very rare. Therefore, this Screening PCR would be useful in small laboratory condition as a screening test for detection of CALR mutations.
Keyword(s): PCR
Session topic: Publication Only
Type: Publication Only
Background
JAK2 and MPL mutation play an important role in diagnosis of myeloid proliferative neoplasm (MPN). Another key marker in the molecular diagnosis of MPNs, mutations in CALR, was recently reported. Known mutations are all located at exon 9 and they are somatic mutations of deletion or insertions; the two commonest abnormalities account for 80-90% of CALR mutations, whereas lesions are highly heterogeneous in the remaining 10-20%.
Aims
Almost previous reports analyzed CALR mutations using Sanger sequencing. However, sequencing method can’t be used easily in every laboratory condition. Authors want to make an easy system to detect CALR mutations which is available in small laboratory without sequencing machine. We investigated the screening PCR to detect CALR mutations in Korean patients with thrombocytosis and compared the results with Sanger sequencing.
Methods
Bone marrow DNA samples were obtained from 83 patients with thrombocytosis. Screening PCR primer sets were designed to detect the wild type (product: 357 bp), type 1 mutation (product: 302 bp) and type 2 mutation (product: 262 bp) in one reaction (Figure 1). And Sanger sequencing was done in hot spot region.
Results
JAK2V617F was detected in 36 patients (33 ET, 1 PMF, 1 MPN, U, and 1 MDS/MPN, U) (Table 1). Two kinds of CALR mutation, type 1 (c.1092_1143del) and type 2 (c.1154_1155insTTGTC) were detected in 16 ET and 1 PMF patients. In ET group, mutational frequencies were 62.3% for JAK2V617F and, 30.2% for CALR, 5.7% for dual positive and 13.2% for dual negative. Among the ET patients without JAK2V617F, frequency of CALR mutation was 80%. The frequency of type 2 CALR mutation (8/16, 50.0%) was higher than type 1 (6/16, 37.5%). Interestingly, one patient with both mutations (1/16, 6.3%) was identified and novel mutation pattern (c.1123_1132delinsTGC) was detected by only Sanger sequencing. Any mutation was not detected in reactive disease group.
Table 1. The frequency of JAK2 and CALR mutations in different patient groups
Disease | ET | PMF | MDS/MPN, U | MPN, U | HES | Reactive | Total |
N=53 | N=3 | N=3 | N=2 | N=1 | N=21 | N=83 | |
JAK2 mutation, N (%) | 33 (62.3%) | 1 (33.3%) | 1 (33.3%) | 1 (50.0%) | 0 (0.0%) | 0 (0.0%) | 36 (43.4%) |
CALR mutation, N (%) | 16 (30.2%)* | 1 (33.3%) | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) | 17 (20.5%) |
Dual positive, N (%) | 3 (5.7%) | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) | 3 (3.6%) |
Dual negative, N (%) | 7 (13.2%) | 1 (33.3%) | 2 (66.7%) | 1 (50.0%) | 1 (100.0%) | 21 (100.0%) | 34 (41.0%) |
CALR mutation (%) | 80% | 50% |
*Type 1: 6 patients (37.5%); Type 2: 8 patients (50.0%); Type 1 & Type 2: 1 patient (6.3%) and novel mutation: 1 patient (6.3%)
Abbreviation: ET; essential thrombocythemia, PMF; primary myelofibrosis, MDS/MPN,U; myelodysplastic syndrome/myeloproliferative neoplasm, unclassifiable, MPN,U; myeloproliferative neoplasm, unclassifiable, HES; hypereosonophilic syndrome, Reactive; reactive disease group with non-hematologic malignancy
Summary
Screening PCR could detect the 94.1% (16/17) of mutation cases, and this method showed the concordant result with Sanger sequencing in the case of type 1 and type 2 mutations. One novel mutation was only identified with Sanger sequencing. Our system can’t detect single base pair substitution or small base insertions like the mutation of this case; however, the frequency of these mutations is very rare. Therefore, this Screening PCR would be useful in small laboratory condition as a screening test for detection of CALR mutations.
Keyword(s): PCR

Session topic: Publication Only


