
Contributions
Type: Publication Only
Background
A frequently occurring chromosomal aberration in acute myeloblastic leukemia (AML) is a translocation involving the mixed lineage leukemia (MLL) gene at 11q23 with one of more than 70 partner genes described to date. Although, in general, MLL rearrangements are related with a poor outcome, there are great differences based only on the translocation partner of MLL. Up to 15% of all MLL rearrangements in AML are due to MLLT10 fusions. MLL-rearrangements result in an upregulation of HOX transcription factor genes. Moreover, EVI1 and MEIS1 upregulation are significantly associated to a poor prognosis in AML and not all MLL-rearranged leukemia cases overexpress these genes.
Aims
In this work we report on the molecular analysis and gene expression profiling for MLL-MLLT10 AML.
Methods
MLL gene fusion was detected by cytogenetics; FISH with LSI MLL dual color, Break Apart probe, and WCP 10 and 11 probes; reverse transcription PCR (RT-PCR) and sequencing. Gene expression was analyzed by real-time PCR in a reaction with specific primers and SybrGreen. Relative quantification was performed by 2-DDCt method. We include analysis of two MLL-MLLT10 harboring leukemias of the FAB-M5 subtype. Both patients obtain complete remission (CR) with positive MRD and disease relapse 4 months after diagnosis. Patient 1 die 3 months after transplantation and second one is currently receiving rescue chemotherapy.
Results
Patient 1 cytogenetic analysis revealed 46,XY,del(10)(p12),add(11)(q23)[20]. FISH analysis using MLL locus specific probe showed a 73% of an uncertain pattern with two nearby split signals. Metaphase FISH with WCP confirmed that chromosome 10 material was present at 11q and that the chromosome 11 material had not been transferred elsewhere in the genome. Patient 2 presented a 46,XX[20], and the same MLL FISH pattern that patient 1. Molecular analysis for the MLL-MLLT10 fusion demonstration was performed based on the presence of NG2 by flow cytometric analysis and a FISH pattern suggesting a gene insertion in 11q23. The cDNA was used in multiplex PCR reactions for detection of the MLL-MLLT10 fusion, and PCR product was sequenced. In patient 1, a mRNA fusion between exon 7 of 5’ MLL mRNA and exon 15 of 3’ MLLT10 mRNA was detected, with frame conservation and the whole sequence of both exons (GenBank ID: KF778381). The breakpoint described here is unusual in 10;11 rearrangements and the same fusion at mRNA level was previously described in only one case. In the second case, the same RT-PCR analysis revealed an mRNA fusion between exon 8 of 5’ MLL mRNA and exon 16 of 3’ MLLT10 mRNA. This MLL-MLLT10 fusion was first described in patient C from Chaplin, et al. 1995.
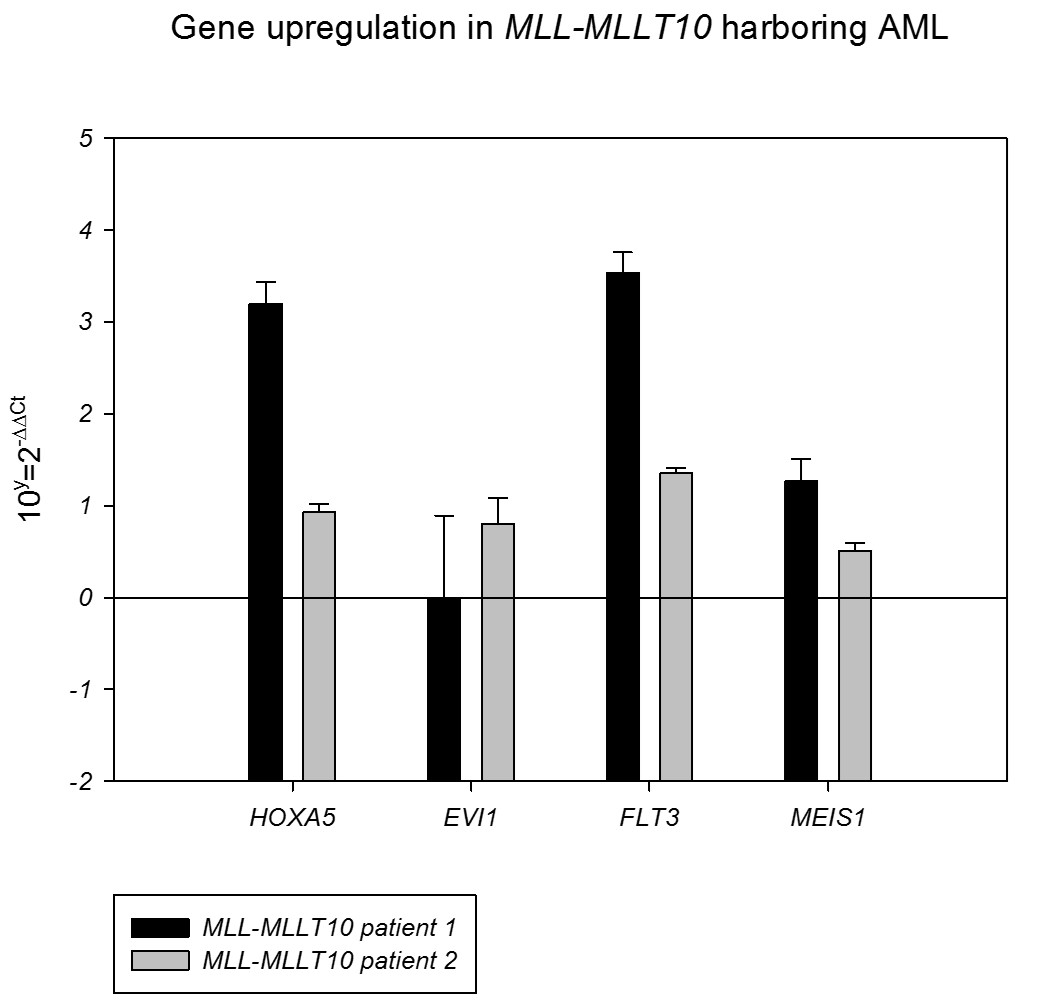
Gene expression analysis showed overexpression of HOXA5, FLT3 and MEIS1 in both cases but EVI1 was only overexpressed in patient 1.
Summary
Molecular studies are required for MLL-MLLT10 detection in cryptic rearrangements and, as illustrated in this work, for the identification of new MLL-MLLT10 fusion points. Gene expression analysis shows overexpression of MEIS1 in both MLL-MLLT10 patients, previously related to poor outcome, and EVI1 only in patient 1.
Keyword(s): MLL
Type: Publication Only
Background
A frequently occurring chromosomal aberration in acute myeloblastic leukemia (AML) is a translocation involving the mixed lineage leukemia (MLL) gene at 11q23 with one of more than 70 partner genes described to date. Although, in general, MLL rearrangements are related with a poor outcome, there are great differences based only on the translocation partner of MLL. Up to 15% of all MLL rearrangements in AML are due to MLLT10 fusions. MLL-rearrangements result in an upregulation of HOX transcription factor genes. Moreover, EVI1 and MEIS1 upregulation are significantly associated to a poor prognosis in AML and not all MLL-rearranged leukemia cases overexpress these genes.
Aims
In this work we report on the molecular analysis and gene expression profiling for MLL-MLLT10 AML.
Methods
MLL gene fusion was detected by cytogenetics; FISH with LSI MLL dual color, Break Apart probe, and WCP 10 and 11 probes; reverse transcription PCR (RT-PCR) and sequencing. Gene expression was analyzed by real-time PCR in a reaction with specific primers and SybrGreen. Relative quantification was performed by 2-DDCt method. We include analysis of two MLL-MLLT10 harboring leukemias of the FAB-M5 subtype. Both patients obtain complete remission (CR) with positive MRD and disease relapse 4 months after diagnosis. Patient 1 die 3 months after transplantation and second one is currently receiving rescue chemotherapy.
Results
Patient 1 cytogenetic analysis revealed 46,XY,del(10)(p12),add(11)(q23)[20]. FISH analysis using MLL locus specific probe showed a 73% of an uncertain pattern with two nearby split signals. Metaphase FISH with WCP confirmed that chromosome 10 material was present at 11q and that the chromosome 11 material had not been transferred elsewhere in the genome. Patient 2 presented a 46,XX[20], and the same MLL FISH pattern that patient 1. Molecular analysis for the MLL-MLLT10 fusion demonstration was performed based on the presence of NG2 by flow cytometric analysis and a FISH pattern suggesting a gene insertion in 11q23. The cDNA was used in multiplex PCR reactions for detection of the MLL-MLLT10 fusion, and PCR product was sequenced. In patient 1, a mRNA fusion between exon 7 of 5’ MLL mRNA and exon 15 of 3’ MLLT10 mRNA was detected, with frame conservation and the whole sequence of both exons (GenBank ID: KF778381). The breakpoint described here is unusual in 10;11 rearrangements and the same fusion at mRNA level was previously described in only one case. In the second case, the same RT-PCR analysis revealed an mRNA fusion between exon 8 of 5’ MLL mRNA and exon 16 of 3’ MLLT10 mRNA. This MLL-MLLT10 fusion was first described in patient C from Chaplin, et al. 1995.
Gene expression analysis showed overexpression of HOXA5, FLT3 and MEIS1 in both cases but EVI1 was only overexpressed in patient 1.
Summary
Molecular studies are required for MLL-MLLT10 detection in cryptic rearrangements and, as illustrated in this work, for the identification of new MLL-MLLT10 fusion points. Gene expression analysis shows overexpression of MEIS1 in both MLL-MLLT10 patients, previously related to poor outcome, and EVI1 only in patient 1.
Keyword(s): MLL



