Lösante Hospital for Children with leukemia,Cancer Research and Genetik Laboratory

Contributions
Type: Publication Only
Background
Acute myeloid leukemia (AML) is a heterogeneous clonal neoplasm characterized by accumulated genetic aberrations, which result in enhanced proliferation, block in differentiation, increased survival of the leukemic blast cells and variable response to therapy. During the past decades, a number of recurrent cytogenetic and molecular genetic abnormalities have been identified in AML such as t(8;21), inv(16), FLT3, NPM1, CEBPA, Ten-Eleven-Translocation 2 (TET2), Kirsten rat sarcoma viral oncogene homolog (KRAS), and Casitas B-cell lymphoma (CBL).
Mutations in TET2 gene could contribute to leukemogenesis by altering epigenetic regulation of transcription via DNA methylation. The incidence of TET2 mutations is approximately 10-20% in AML. RAS mutations, especially KRAS, represent about 90% of cancer-associated mutations. RAS proteins play a major role in cell signaling pathway of cell proliferation, differentiation, and survival. KRAS mutations are the most frequently seen and found in 10-15% of these patients. CBL is a mammalian gene encoding the protein CBL which is an E3 ubiquitin-protein ligase involved in cell signaling and protein ubiquitination. These mutations have also been observed in 1% of AML.
Aims
In this study, we aimed to screen whole TET2, (KRAS) and (CBL) genes by using Next generation sequencing (NGS) analysis to find new possible genetic markers in children with AML.
Methods
Study population consisted of eight patients aged between 1 and 15 years who were admitted to Lösante Hospital for Children with leukemia with the diagnosis of AML. The Patient characteristics of the cases are shown in Table 1. Blood samples were collected with EDTA-containing tubes and DNA was extracted from peripheral blood and bone marrow leukocytes with MagNA Pure automatic DNA isolation instrument (Roche Diagnostics, Manheim, Germany). We utilized NGS to study three candidate genes at TET2, KRAS and CBL. All coding exons of TET2 (exons 3 and 11) were presented by 27 amplicons. Beside, two primer pairs were amplified known mutational hotspot regions to describe the RING finger domain and linker sequence for CBL (exons 8 and 9) and KRAS (exons 2 and 3).
Results
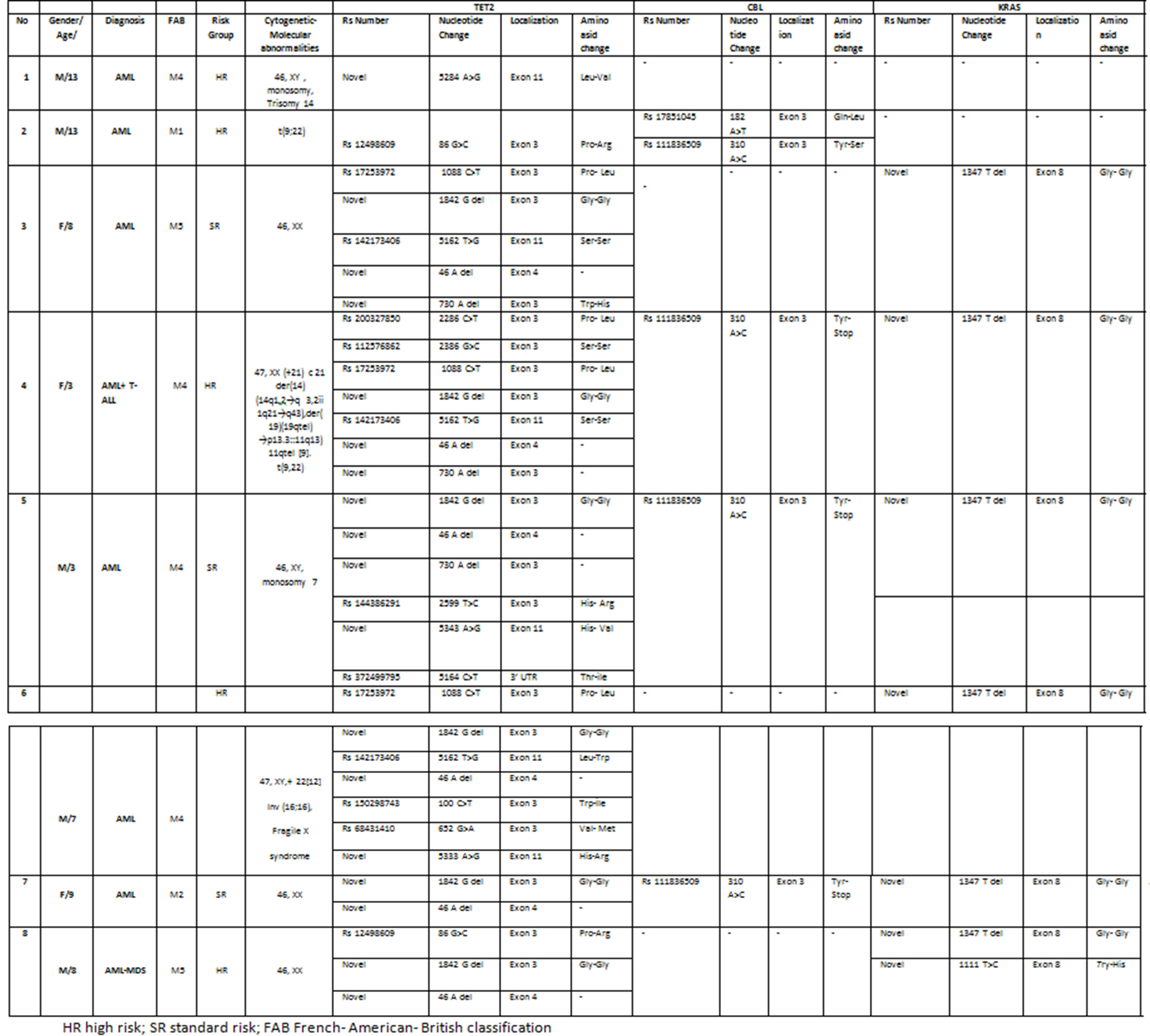
We detected 16 variants in TET2, 2 variants both KRAS and CBL. The most of TET2 variants were described in the largest exon 3 and 11; 1842 G>- (6/8 patients, 75%) and 1088 C>T (5/8, 62,5%) variants, 5162 T>G(5/8, 62,5%) in exon 3 and exon 11 of TET2, respectively. 310 A>C (7/8 87,5%) variation was the highest among the variants in intron 2 of KRAS. 1347 T>- in exon 8 of CBL was detected 6/8 patients (75%).Table 1 summarizes the association of patients’ characteristics and variants of TET2, KRAS and CBL profiles.
Table 1. Demographic characteristics and TET2, KRAS, and CBL variants of children with AML
Summary
In this study, we screened the mutations of TET2, KRAS and CBL genes in pediatric AML patients. We used an amplicon based sequencing method to find possible new genetic markers for leukemia diagnosis. TET2, KRAS and CBL genes were selected based on recent studies on genetic abnormalities in AML and other hematologic malignancies. In 8 patients, we report novel mutations at TET2 and CBL genes. Seven of 16 substitutions were missense mutations in the exon and UTR regions. These mutations may result in truncated translation of protein.
In conclusion, we found that TET2 mutations are more frequent than KRAS, and CBL mutations in pediatric AML in this study. The usage of NGS to search for TET2, KRAS, and CBL mutations might be fruitful, however, these results need to be confirmed by further studies on a larger number of patients.
Keyword(s): AML, Mutation, Pediatric
Type: Publication Only
Background
Acute myeloid leukemia (AML) is a heterogeneous clonal neoplasm characterized by accumulated genetic aberrations, which result in enhanced proliferation, block in differentiation, increased survival of the leukemic blast cells and variable response to therapy. During the past decades, a number of recurrent cytogenetic and molecular genetic abnormalities have been identified in AML such as t(8;21), inv(16), FLT3, NPM1, CEBPA, Ten-Eleven-Translocation 2 (TET2), Kirsten rat sarcoma viral oncogene homolog (KRAS), and Casitas B-cell lymphoma (CBL).
Mutations in TET2 gene could contribute to leukemogenesis by altering epigenetic regulation of transcription via DNA methylation. The incidence of TET2 mutations is approximately 10-20% in AML. RAS mutations, especially KRAS, represent about 90% of cancer-associated mutations. RAS proteins play a major role in cell signaling pathway of cell proliferation, differentiation, and survival. KRAS mutations are the most frequently seen and found in 10-15% of these patients. CBL is a mammalian gene encoding the protein CBL which is an E3 ubiquitin-protein ligase involved in cell signaling and protein ubiquitination. These mutations have also been observed in 1% of AML.
Aims
In this study, we aimed to screen whole TET2, (KRAS) and (CBL) genes by using Next generation sequencing (NGS) analysis to find new possible genetic markers in children with AML.
Methods
Study population consisted of eight patients aged between 1 and 15 years who were admitted to Lösante Hospital for Children with leukemia with the diagnosis of AML. The Patient characteristics of the cases are shown in Table 1. Blood samples were collected with EDTA-containing tubes and DNA was extracted from peripheral blood and bone marrow leukocytes with MagNA Pure automatic DNA isolation instrument (Roche Diagnostics, Manheim, Germany). We utilized NGS to study three candidate genes at TET2, KRAS and CBL. All coding exons of TET2 (exons 3 and 11) were presented by 27 amplicons. Beside, two primer pairs were amplified known mutational hotspot regions to describe the RING finger domain and linker sequence for CBL (exons 8 and 9) and KRAS (exons 2 and 3).
Results
We detected 16 variants in TET2, 2 variants both KRAS and CBL. The most of TET2 variants were described in the largest exon 3 and 11; 1842 G>- (6/8 patients, 75%) and 1088 C>T (5/8, 62,5%) variants, 5162 T>G(5/8, 62,5%) in exon 3 and exon 11 of TET2, respectively. 310 A>C (7/8 87,5%) variation was the highest among the variants in intron 2 of KRAS. 1347 T>- in exon 8 of CBL was detected 6/8 patients (75%).Table 1 summarizes the association of patients’ characteristics and variants of TET2, KRAS and CBL profiles.
Table 1. Demographic characteristics and TET2, KRAS, and CBL variants of children with AML
Summary
In this study, we screened the mutations of TET2, KRAS and CBL genes in pediatric AML patients. We used an amplicon based sequencing method to find possible new genetic markers for leukemia diagnosis. TET2, KRAS and CBL genes were selected based on recent studies on genetic abnormalities in AML and other hematologic malignancies. In 8 patients, we report novel mutations at TET2 and CBL genes. Seven of 16 substitutions were missense mutations in the exon and UTR regions. These mutations may result in truncated translation of protein.
In conclusion, we found that TET2 mutations are more frequent than KRAS, and CBL mutations in pediatric AML in this study. The usage of NGS to search for TET2, KRAS, and CBL mutations might be fruitful, however, these results need to be confirmed by further studies on a larger number of patients.
Keyword(s): AML, Mutation, Pediatric



