Department of Haematology

Contributions
Type: Publication Only
Background
It has formerly been suggested that the high incidence of cancer in human, as compared to non-human primates, could likely be related to mal-adaptations during the recent evolutionary past. MYEOV, an oncogene in multiple myeloma (MM), was recently characterized as a human-specific protein-coding gene of de novo origin from long non-coding RNA. In 2010, MYEOV expression was documented in primary MM cells and in purified plasma cells from patients with monoclonal gammopathy of undetermined significance however was scarcely detected in bone marrow plasma cells and was absent in normal plasmablasts or memory B cells. In addition, the gene was identified as causally involved in promoting MM cells propagation. MYEOV has the potential to encode for two protein isoforms, namely for a 313 amino acid peptide (MYEOV-313) and for a shorter one (MYEOV-255). Solid Western-blot assays support the production of both the proteins while MYEOV-313 expression has been validated in patients with MM. Both the peptides seem to be directed to the membrane nonetheless are of, yet, unknown function.
Aims
Illuminate, via in silico analysis, the evolutionary path leading to MYEOV. Highlight that a captivating exaptation event of an ancient transposable element (TE) significantly contributed to the creation of MYEOV gene structure.
Methods
BLASTN search against the GenBank human genomic database and BLASTP search against the GenBank nonredundant protein database were used to exclude MYEOV origination via gene duplication and to verify unambiguous MYEOV orthologs. MYEOV orthologs annotated in the respective databases from NCBI and Ensembl were identified by searching with the gene name. Whole-genome shotgun (WGS) sequence contigs, including within the DNA segments used to curate the reference genomic sequences of MYEOV orthologs, were downloaded from the NCBI Gene database. The WGS sequence contigs were used to build MYEOV locus alignment blocks between human and the 11 species with annotated orthologs, via BLASTN. Each alignment block was manually scrutinized for validating the primary open reading frame (ORF) of the annotated orthologs. Search for MYEOV upstream ORFs was performed with the ORF Finder program. MYEOV splice site analysis was performed with the Human Splicing Finder Version 2.4.1 program. MYEOV was scanned for the presence of TEs by Repeat Masker. MYEOV locus syntenic alignments of numerous Vertebrates were downloaded from the UCSC Genome Browser Database (UCSC GBD). The automated alignments were used to extract syntenic DNA segments in numerous mammals that flank MYEOV-313 start codon. The DNA segments extracted were scanned by RepeatMasker.
Results
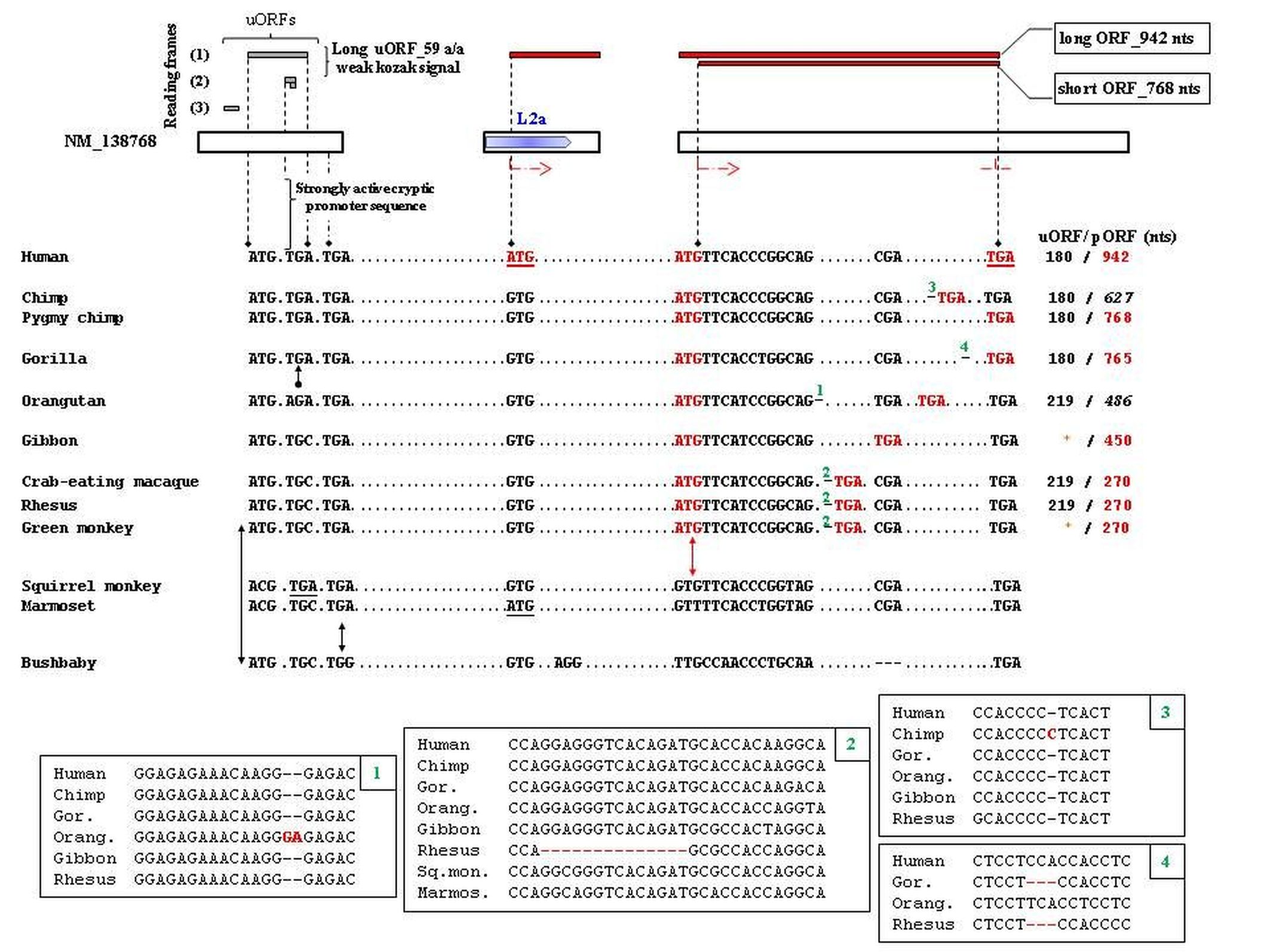
MYEOV BLASTN search yields no significant similarity with any other coding sequence in the human genome. Syntenic alignments, extracted from the UCSC GBD, signify that the DNA segment where MYEOV locates emerged in Eutherian mammals. An important mutational event occurred in Catarrhini resulting in the de novo gain of a translatable ORF in a genomic region that seems noncoding in other species. That is the acquisition of a MYEOV-255 start codon. During evolution in the Catarrhini genome, MYEOV acquired a gradually elongated translatable ORF, a gradually shortened translation-regulatory upstream ORF, as well as introns and alternatively spliced mRNA variants. During Homo / Pan separation time, a momentous point mutation was introduced in human resulting in the acquisition of MYEOV-313 start codon. Accordingly, many of the automated annotations of MYEOV orthologs, provided by the NCBI and Ensembl automated pipelines, contain inaccuracies. MYEOV-313 start codon is included in a genomic region reported from RepeatMasker to match a TE of the L2 family. The RepeatMasker match appears degenerated however in-depth phylogenetic analysis verified the presence of the L2 repeat in syntenic DNA segments in many mammals.
Summary
MYEOV is a Primate Orphan Gene. MYEOV-255 is conserved in Hominines while MYEOV-313 represents a human-specific peptide. MYEOV was characterized as a human-specific de novo gene because the existence of two protein isoforms was not specifically addressed. MYEOV-313 start codon was evolutionary provided by a L2 TE. Could this rare exaptation event represent a human mal-adaptation, is a tantalizing possibility. Accordingly, MYEOV-313 biological role warrants further research, especially because targeting a human-specific peptide could theoretically cause less adverse effects than targeting components of evolutionary conserved signaling cascades.
Keyword(s): Multiple myeloma, Oncogene
Type: Publication Only
Background
It has formerly been suggested that the high incidence of cancer in human, as compared to non-human primates, could likely be related to mal-adaptations during the recent evolutionary past. MYEOV, an oncogene in multiple myeloma (MM), was recently characterized as a human-specific protein-coding gene of de novo origin from long non-coding RNA. In 2010, MYEOV expression was documented in primary MM cells and in purified plasma cells from patients with monoclonal gammopathy of undetermined significance however was scarcely detected in bone marrow plasma cells and was absent in normal plasmablasts or memory B cells. In addition, the gene was identified as causally involved in promoting MM cells propagation. MYEOV has the potential to encode for two protein isoforms, namely for a 313 amino acid peptide (MYEOV-313) and for a shorter one (MYEOV-255). Solid Western-blot assays support the production of both the proteins while MYEOV-313 expression has been validated in patients with MM. Both the peptides seem to be directed to the membrane nonetheless are of, yet, unknown function.
Aims
Illuminate, via in silico analysis, the evolutionary path leading to MYEOV. Highlight that a captivating exaptation event of an ancient transposable element (TE) significantly contributed to the creation of MYEOV gene structure.
Methods
BLASTN search against the GenBank human genomic database and BLASTP search against the GenBank nonredundant protein database were used to exclude MYEOV origination via gene duplication and to verify unambiguous MYEOV orthologs. MYEOV orthologs annotated in the respective databases from NCBI and Ensembl were identified by searching with the gene name. Whole-genome shotgun (WGS) sequence contigs, including within the DNA segments used to curate the reference genomic sequences of MYEOV orthologs, were downloaded from the NCBI Gene database. The WGS sequence contigs were used to build MYEOV locus alignment blocks between human and the 11 species with annotated orthologs, via BLASTN. Each alignment block was manually scrutinized for validating the primary open reading frame (ORF) of the annotated orthologs. Search for MYEOV upstream ORFs was performed with the ORF Finder program. MYEOV splice site analysis was performed with the Human Splicing Finder Version 2.4.1 program. MYEOV was scanned for the presence of TEs by Repeat Masker. MYEOV locus syntenic alignments of numerous Vertebrates were downloaded from the UCSC Genome Browser Database (UCSC GBD). The automated alignments were used to extract syntenic DNA segments in numerous mammals that flank MYEOV-313 start codon. The DNA segments extracted were scanned by RepeatMasker.
Results
MYEOV BLASTN search yields no significant similarity with any other coding sequence in the human genome. Syntenic alignments, extracted from the UCSC GBD, signify that the DNA segment where MYEOV locates emerged in Eutherian mammals. An important mutational event occurred in Catarrhini resulting in the de novo gain of a translatable ORF in a genomic region that seems noncoding in other species. That is the acquisition of a MYEOV-255 start codon. During evolution in the Catarrhini genome, MYEOV acquired a gradually elongated translatable ORF, a gradually shortened translation-regulatory upstream ORF, as well as introns and alternatively spliced mRNA variants. During Homo / Pan separation time, a momentous point mutation was introduced in human resulting in the acquisition of MYEOV-313 start codon. Accordingly, many of the automated annotations of MYEOV orthologs, provided by the NCBI and Ensembl automated pipelines, contain inaccuracies. MYEOV-313 start codon is included in a genomic region reported from RepeatMasker to match a TE of the L2 family. The RepeatMasker match appears degenerated however in-depth phylogenetic analysis verified the presence of the L2 repeat in syntenic DNA segments in many mammals.
Summary
MYEOV is a Primate Orphan Gene. MYEOV-255 is conserved in Hominines while MYEOV-313 represents a human-specific peptide. MYEOV was characterized as a human-specific de novo gene because the existence of two protein isoforms was not specifically addressed. MYEOV-313 start codon was evolutionary provided by a L2 TE. Could this rare exaptation event represent a human mal-adaptation, is a tantalizing possibility. Accordingly, MYEOV-313 biological role warrants further research, especially because targeting a human-specific peptide could theoretically cause less adverse effects than targeting components of evolutionary conserved signaling cascades.
Keyword(s): Multiple myeloma, Oncogene



