
Contributions
Abstract: S161
Type: Oral Presentation
Session title: Correlating mutations with phenotype in MDS
Background
Myelodysplastic syndromes (MDS) lie on a continuum of myeloid neoplastic progression between clonal hematopoiesis of indeterminate potential (CHIP) and myeloid leukemia. Innate immune signalling in the marrow has been identified as a determinant of progression, particularly in low-risk (LR)-MDS, suggesting that it may be possible to prevent or delay progression by suitably targeted anti-inflammatory therapy. However, while early data point to a pivotal role for the NLRP3-inflammasome, little is known about the extent or functional relevance of patient-specific variations in inflammatory network activity.
Aims
Our objective was to assess the diversity of pre-treatment bone marrow inflammation in LR-MDS compared to non-CHIP, CHIP, high-risk (HR)-MDS and CMML patients and to investigate associations with phenotype, genotype and outcome in LR-MDS.
Methods
We analyzed a total of 102 bone marrow samples (15 non-CHIP; 12 CHIP; 47 LR-MDS; 14 HR-MDS; 14 CMML) collected under informed consent as part of the MDS registry and the BoHemE Study (NCT02867085) at the University Hospitals in Dresden and Leipzig. BM-MNC RNA was analyzed by qPCR TaqMan® Array for the expression of 12 genes encoding inflammasome-related proteins (NLRP3, PYCARD, CASP1, IL1B, IL18, S100A9, NLRP1, NLRC4, AIM2, CASP3, CASP4, CASP5). In parallel, we measured the levels of 14 circulating inflammatory mediators (e.g. IL1B, IL18, S100A9) in paired plasma samples by LuminexTM and conventional ELISA. Inflammasome data were then correlated with mutation status and clinical parameters.
Results
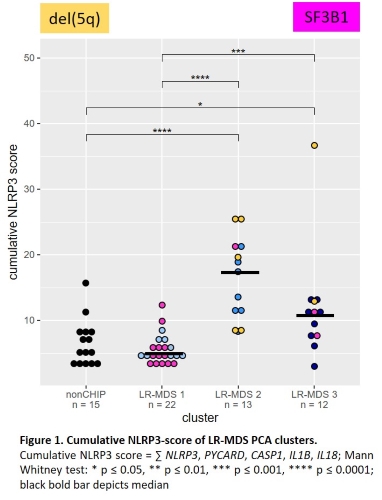
Inflammasome gene expression was generally higher in LR-MDS than in either CHIP or HR-MDS. In LR-MDS samples selected inflammatory biomarkers such as NLRP3, IL1B, IL18, and S100A9 were differentially expressed and correlated, indicating the presence of distinct patterns of inflammation within this cohort. A principal component analysis (PCA) performed in order to identify consistent subgroups in the gene expression separated the LR-MDS cohort into three defined clusters based predominantly on the levels of PYCARD, IL18, S100A9 and NLRC4. Re-examination of these groups for NLRP3-related gene expression confirmed low, medium and high levels (Figure 1).
Low NLRP3-inflammasome state (LR-MDS 1) associated with the presence of SF3B1 mutation/ring sideroblasts, hypercellular bone marrow, transfusion dependence and expanded erythropoiesis, as well as low IL1B mRNA and protein expression. High NLRP3-inflammasome state (LR-MDS 2) associated with del(5q), hypocellular bone marrow and transfusion independence, as well as high IL1B mRNA and protein expression. Medium NLRP3-inflammasome state (LR-MDS 3) associated with fewer mutations, hypocellular bone marrow and expanded erythropoiesis, as well as high IL1B mRNA and protein expression.
Conclusion
We demonstrate the presence of distinct inflammasome activation states in LR-MDS and identify associated biomarkers (IL1B, SF3B1mut, del(5q)) of potential relevance for patient stratification to anti-inflammatory therapies. We are currently analyzing gene expression in sorted cell populations to further increase the resolution of our analysis, which will be validated in the prospective CANFIRE trial investigating the IL1B antibody canakinumab in LR-MDS.
Keyword(s): Bone Marrow, Bone marrow failure, Inflammation, MDS
Abstract: S161
Type: Oral Presentation
Session title: Correlating mutations with phenotype in MDS
Background
Myelodysplastic syndromes (MDS) lie on a continuum of myeloid neoplastic progression between clonal hematopoiesis of indeterminate potential (CHIP) and myeloid leukemia. Innate immune signalling in the marrow has been identified as a determinant of progression, particularly in low-risk (LR)-MDS, suggesting that it may be possible to prevent or delay progression by suitably targeted anti-inflammatory therapy. However, while early data point to a pivotal role for the NLRP3-inflammasome, little is known about the extent or functional relevance of patient-specific variations in inflammatory network activity.
Aims
Our objective was to assess the diversity of pre-treatment bone marrow inflammation in LR-MDS compared to non-CHIP, CHIP, high-risk (HR)-MDS and CMML patients and to investigate associations with phenotype, genotype and outcome in LR-MDS.
Methods
We analyzed a total of 102 bone marrow samples (15 non-CHIP; 12 CHIP; 47 LR-MDS; 14 HR-MDS; 14 CMML) collected under informed consent as part of the MDS registry and the BoHemE Study (NCT02867085) at the University Hospitals in Dresden and Leipzig. BM-MNC RNA was analyzed by qPCR TaqMan® Array for the expression of 12 genes encoding inflammasome-related proteins (NLRP3, PYCARD, CASP1, IL1B, IL18, S100A9, NLRP1, NLRC4, AIM2, CASP3, CASP4, CASP5). In parallel, we measured the levels of 14 circulating inflammatory mediators (e.g. IL1B, IL18, S100A9) in paired plasma samples by LuminexTM and conventional ELISA. Inflammasome data were then correlated with mutation status and clinical parameters.
Results
Inflammasome gene expression was generally higher in LR-MDS than in either CHIP or HR-MDS. In LR-MDS samples selected inflammatory biomarkers such as NLRP3, IL1B, IL18, and S100A9 were differentially expressed and correlated, indicating the presence of distinct patterns of inflammation within this cohort. A principal component analysis (PCA) performed in order to identify consistent subgroups in the gene expression separated the LR-MDS cohort into three defined clusters based predominantly on the levels of PYCARD, IL18, S100A9 and NLRC4. Re-examination of these groups for NLRP3-related gene expression confirmed low, medium and high levels (Figure 1).
Low NLRP3-inflammasome state (LR-MDS 1) associated with the presence of SF3B1 mutation/ring sideroblasts, hypercellular bone marrow, transfusion dependence and expanded erythropoiesis, as well as low IL1B mRNA and protein expression. High NLRP3-inflammasome state (LR-MDS 2) associated with del(5q), hypocellular bone marrow and transfusion independence, as well as high IL1B mRNA and protein expression. Medium NLRP3-inflammasome state (LR-MDS 3) associated with fewer mutations, hypocellular bone marrow and expanded erythropoiesis, as well as high IL1B mRNA and protein expression.

Conclusion
We demonstrate the presence of distinct inflammasome activation states in LR-MDS and identify associated biomarkers (IL1B, SF3B1mut, del(5q)) of potential relevance for patient stratification to anti-inflammatory therapies. We are currently analyzing gene expression in sorted cell populations to further increase the resolution of our analysis, which will be validated in the prospective CANFIRE trial investigating the IL1B antibody canakinumab in LR-MDS.
Keyword(s): Bone Marrow, Bone marrow failure, Inflammation, MDS


